Note
Welcome to our latest work: CareLlama (Care Alpaca), a medical large language model that combines dozens of publicly available medical fine-tuning datasets and openly available medical large language models to promote the rapid development of medical LLM: https://github.com/WangRongsheng/CareLlama
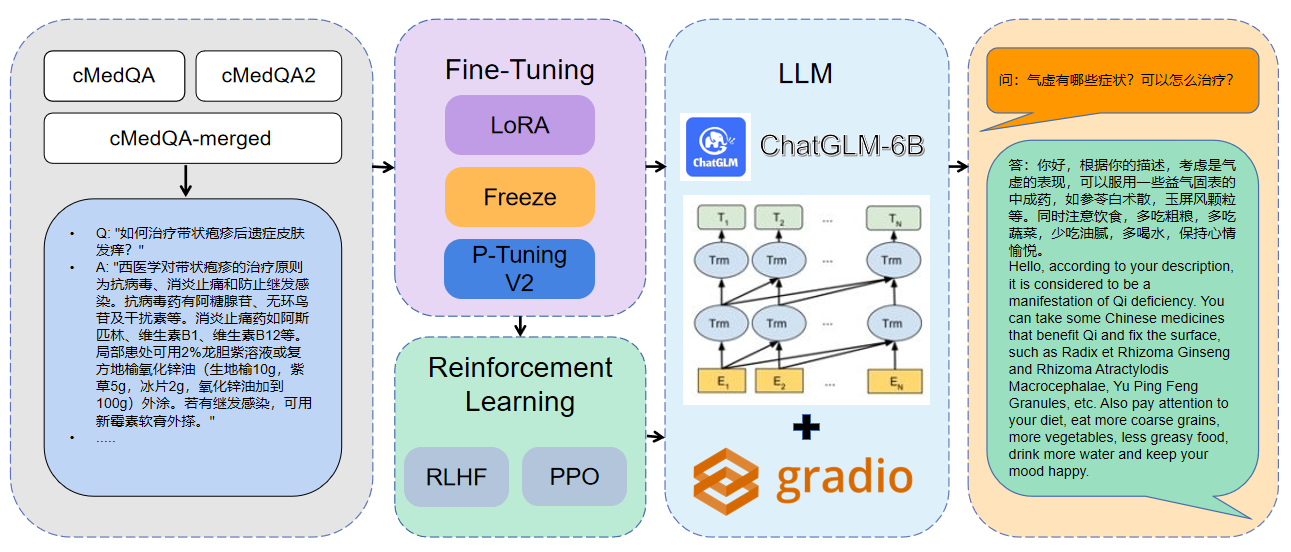
1 The data used is cMedQA2
| project | Dataset | Base model |
|---|---|---|
| ChatMed | Consult contains 50w + online consultation + ChatGPT reply, TCM traditional Chinese medicine diagnosis and treatment data set has not been published | LLaMA-7B |
| ChatDoctor | HealthCareMagic-100k contains 100k+ real patient-doctor dialogue dataset, icliniq-10k contains 10k+ patient-doctor dialogue dataset, GenMedGPT-5k contains 5k+ doctor-patient dialogue dataset generated by GPT | LLaMA-7B |
| Med-ChatGLM | Huatuo-data, Huatuo-liver-cancer | ChatGLM-6B |
| Huatuo-Llama-Med-Chinese | Huatuo-data, Huatuo-liver-cancer | LLaMA-7B |
| DoctorGLM | CMD., MedDialog, ChatDoctor project dataset | ChatGLM-6B |
| MedicalGPT-zh | Data not open source | ChatGLM-6B |
| Dr.LLaMA | LLaMA | |
| Medical_NLP 2 | - | - |
| CMCQA 3 | - | - |
| QiZhenGPT | - | - |
| LLM-Pretrain-FineTune | - | - |
| PMC-LLaMA | - | LLaMA-7B |
| BianQue | - | - |
| medAlpaca | - | LLaMA-7B |
| MedicalGPT | - | - |
| LLM-Pretrain-FineTune | - | - |
| ShenNong-TCM-LLM | - | - |
| Sunsimiao | - | - |
| CMLM-ZhongJing | - | - |
| ZhongJing | - | - |
| Ming | - | - |
| DISC-MedLLM | - | - |
pip install - r requirements . txt CUDA_VISIBLE_DEVICES = 0 python MedQA - ChatGLM / finetune . py
- - do_train
- - dataset merged - cMedQA
- - finetuning_type lora
- - output_dir . / med - lora
- - per_device_train_batch_size 32
- - gradient_accumulation_steps 256
- - lr_scheduler_type cosine
- - logging_steps 500
- - save_steps 1000
- - learning_rate 5e-5
- - num_train_epochs 10.0
- - fp16 CUDA_VISIBLE_DEVICES = 0 python MedQA - ChatGLM / finetune . py
- - do_train
- - dataset merged - cMedQA
- - finetuning_type freeze
- - output_dir . / med - freeze
- - per_device_train_batch_size 32
- - gradient_accumulation_steps 256
- - lr_scheduler_type cosine
- - logging_steps 500
- - save_steps 1000
- - learning_rate 5e-5
- - num_train_epochs 10.0
- - fp16 CUDA_VISIBLE_DEVICES = 1 python MedQA - ChatGLM / finetune . py
- - do_train - - dataset merged - cMedQA
- - finetuning_type p_tuning
- - output_dir . / med - p_tuning
- - per_device_train_batch_size 32
- - gradient_accumulation_steps 256
- - lr_scheduler_type cosine
- - logging_steps 500
- - save_steps 1000
- - learning_rate 5e-5
- - num_train_epochs 10.0
- - fp16For more parameter information, you can view the detailed explanation of docs/parameters.md.
Multi-GPU distributed training:
# 配置分布式参数
accelerate config
# 分布式训练
accelerate launch src / finetune . py
- - do_train
- - dataset Huatuo , CMD , MedDialog , guanaco , cognition
- - finetuning_type lora
- - output_dir med - lora
- - per_device_train_batch_size 16
- - gradient_accumulation_steps 4
- - lr_scheduler_type cosine
- - logging_steps 10
- - save_steps 1000
- - learning_rate 5e-5
- - num_train_epochs 3.0
- - fp16
- - ddp_find_unused_parameters False # 分布式训练时,LoRA微调需要添加防止报错
- - plot_loss CUDA_VISIBLE_DEVICES = 0 python MedQA - ChatGLM / web_demo . py
- - checkpoint_dir med - lora /
( med - freez / )
( med - p_tuning / ) CUDA_VISIBLE_DEVICES = 0 python MedQA - ChatGLM / infer . py
- - checkpoint_dir med - lora /
( med - freez / )
( med - p_tuning / )Merge the model:
CUDA_VISIBLE_DEVICES = 0 python MedQA - ChatGLM / export_weights . py
- - finetuning_weights_path . / med - lora
- - save_weights_path . / save_loraLoading the merged model:
CUDA_VISIBLE_DEVICES = 0 python MedQA - ChatGLM / load_export_weights . py
- - save_weights_path . / save_lora| Fine-tuning method | Model weight | Training duration | Training rounds |
|---|---|---|---|
| LoRA | MedQA-ChatGLM-LoRA | 28h | 10 |
| P-Tuning V2 | MedQA-ChatGLM-PTuningV2 | 27h | 10 |
| Freeze | MedQA-ChatGLM-Freeze | 28h | 10 |
* The experiment was conducted on Linux system, A100 (1X, 80GB)
The resources related to this project are for academic research only and are strictly prohibited for commercial purposes. When using parts involving third-party code, please strictly follow the corresponding open source protocol. The content generated by the model is affected by factors such as model calculation, randomness and quantitative accuracy losses, and this project cannot guarantee its accuracy. Most of the data sets of this project are generated by models and cannot be used as the basis for actual medical diagnosis even if they comply with certain medical facts. This project assumes no legal liability for any content output by the model, nor is it liable for any losses that may arise from the use of relevant resources and output results.