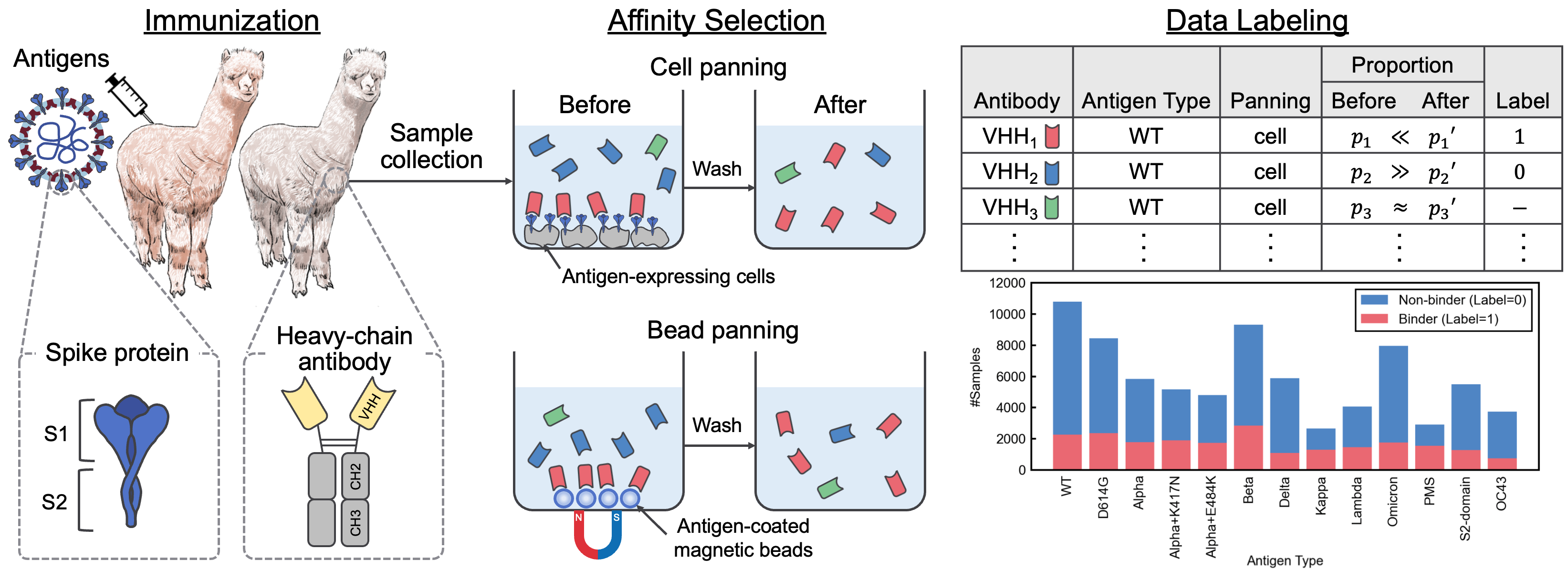
This repository contains the supplementary material accompanying the paper "A SARS-CoV-2 Interaction Dataset and VHH Sequence Corpus for Antibody Language Models." In this paper, we introduced AVIDa-SARS-CoV-2, a labeled dataset of SARS-CoV-2-VHH interactions, and VHHCorpus-2M, which contains over two million VHH sequences, providing novel datasets for the evaluation and pre-training of antibody language models. The datasets are available at https://datasets.cognanous.com under a CC BY-NC 4.0 license.
To get started, clone this repository and run the following command to create a virtual environment.
python -m venv ./venv
source ./venv/bin/activate
pip install -r requirements.txt| Dataset | Links |
|---|---|
| VHHCorpus-2M | Hugging Face Hub Project Page |
| AVIDa-SARS-CoV-2 | Hugging Face Hub Project Page |
The code for converting the raw data (FASTQ file) obtained from next-generation sequencing (NGS) into the labeled dataset, AVIDa-SARS-CoV-2, can be found under ./dataset.
We released the FASTQ files for antigen type "OC43" here so that the data processing can be reproduced.
First, you need to create a Docker image.
docker build -t vhh_constructor:latest ./dataset/vhh_constructorAfter placing the FASTQ files under dataset/raw/fastq, execute the following command to output a labeled CSV file.
bash ./dataset/preprocess.shVHHBERT is a RoBERTa-based model pre-trained on two million VHH sequences in VHHCorpus-2M. VHHBERT can be pre-trained with the following commands.
python benchmarks/pretrain.py --vocab-file "benchmarks/data/vocab_vhhbert.txt"
--epochs 20
--batch-size 128
--save-dir "outputs"Arguments:
| Argument | Required | Default | Description |
|---|---|---|---|
| --vocab-file | Yes | Path of the vocabulary file | |
| --epochs | No | 20 | Number of epochs |
| --batch-size | No | 128 | Size of mini-batch |
| --seed | No | 123 | Random seed |
| --save-dir | No | ./saved | Path of the save directory |
The pre-trained VHHBERT, released under the MIT License, is available on the Hugging Face Hub.
To evaluate the performance of various pre-trained language models for antibody discovery, we defined a binary classification task to predict the binding or non-binding of unknown antibodies to 13 antigens using AVIDa-SARS-CoV-2. For more information on the benchmarking task, see the paper.
Fine-tuning of the language models can be performed using the following command.
python benchmarks/finetune.py --palm-type "VHHBERT"
--epochs 30
--batch-size 32
--save-dir "outputs"palm-type must be one of the following:
VHHBERTVHHBERT-w/o-PTAbLangAntiBERTa2AntiBERTa2-CSSPIgBertProtBertESM-2-150MESM-2-650MArguments:
| Argument | Required | Default | Description |
|---|---|---|---|
| --palm-type | No | VHHBERT | Model name |
| --embeddings-file | No | ./benchmarks/data/antigen_embeddings.pkl | Path of embeddings file for antigens |
| --epochs | No | 20 | Number of epochs |
| --batch-size | No | 128 | Size of mini-batch |
| --seed | No | 123 | Random seed |
| --save-dir | No | ./saved | Path of the save directory |
If you use AVIDa-SARS-CoV-2, VHHCorpus-2M, or VHHBERT in your research, please use the following citation.
@inproceedings{tsuruta2024sars,
title={A {SARS}-{C}o{V}-2 Interaction Dataset and {VHH} Sequence Corpus for Antibody Language Models},
author={Hirofumi Tsuruta and Hiroyuki Yamazaki and Ryota Maeda and Ryotaro Tamura and Akihiro Imura},
booktitle={Advances in Neural Information Processing Systems 37},
year={2024}
}