Sklearn genetic opt
0.11.1


Scikit-Learn使用進化算法模型的超參數調整和特徵選擇。
這本來可以替代Scikit-Learn中流行方法的一種替代方法,例如網格搜索和對超參數調整的隨機網格搜索,以及從RFE(遞歸功能消除),從模型中選擇以進行特徵選擇。
Sklearn-Genetic-Opt使用來自DEAP(Python中的分佈式進化算法)的進化算法來選擇優化(最大或最小)的一組超參數,可以將交叉驗證得分用於回歸和分類問題。
文檔可在此處提供
可視化培訓的進度:
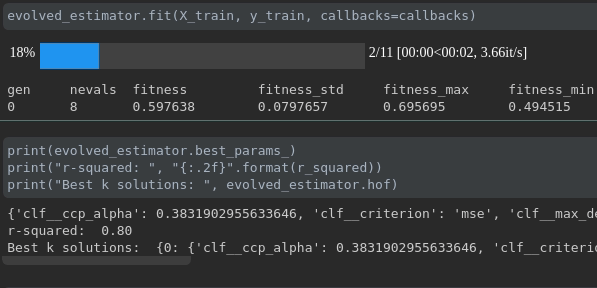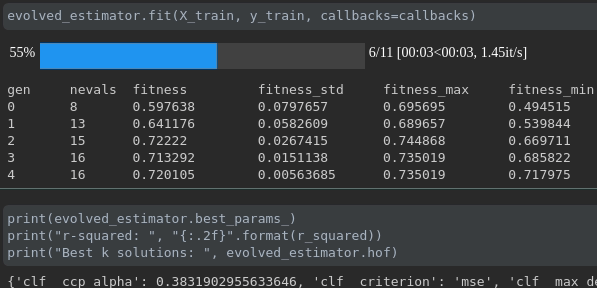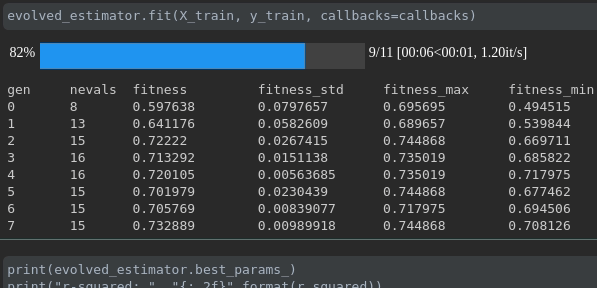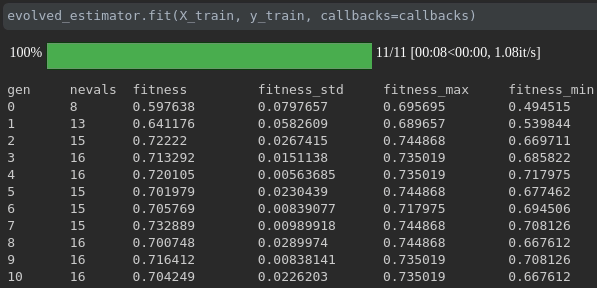
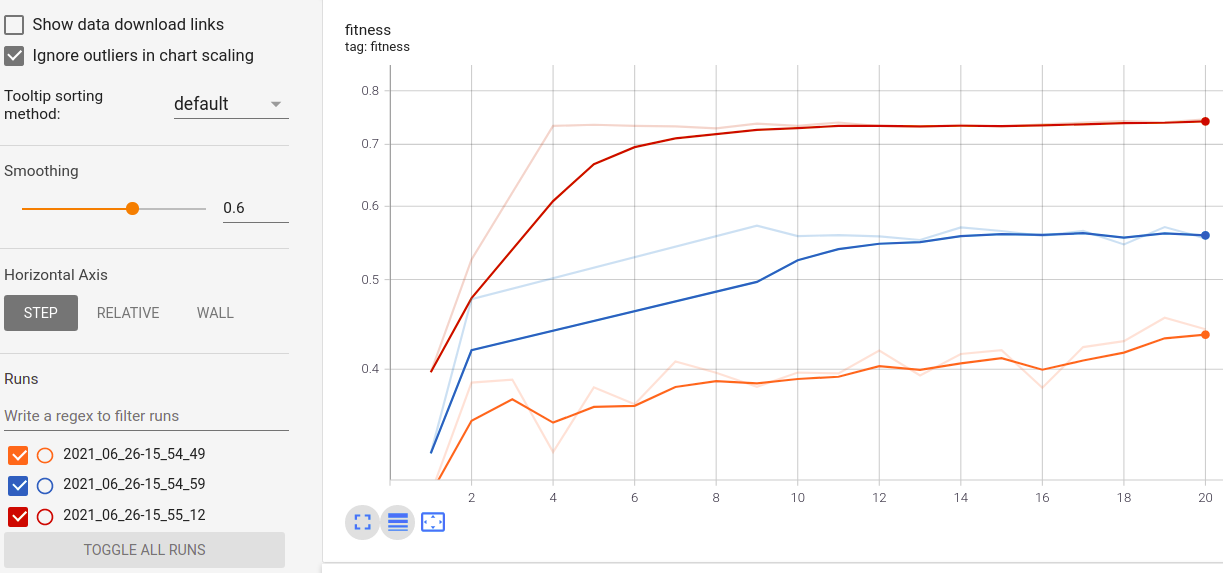
實時指標可視化和跨行駛的比較:
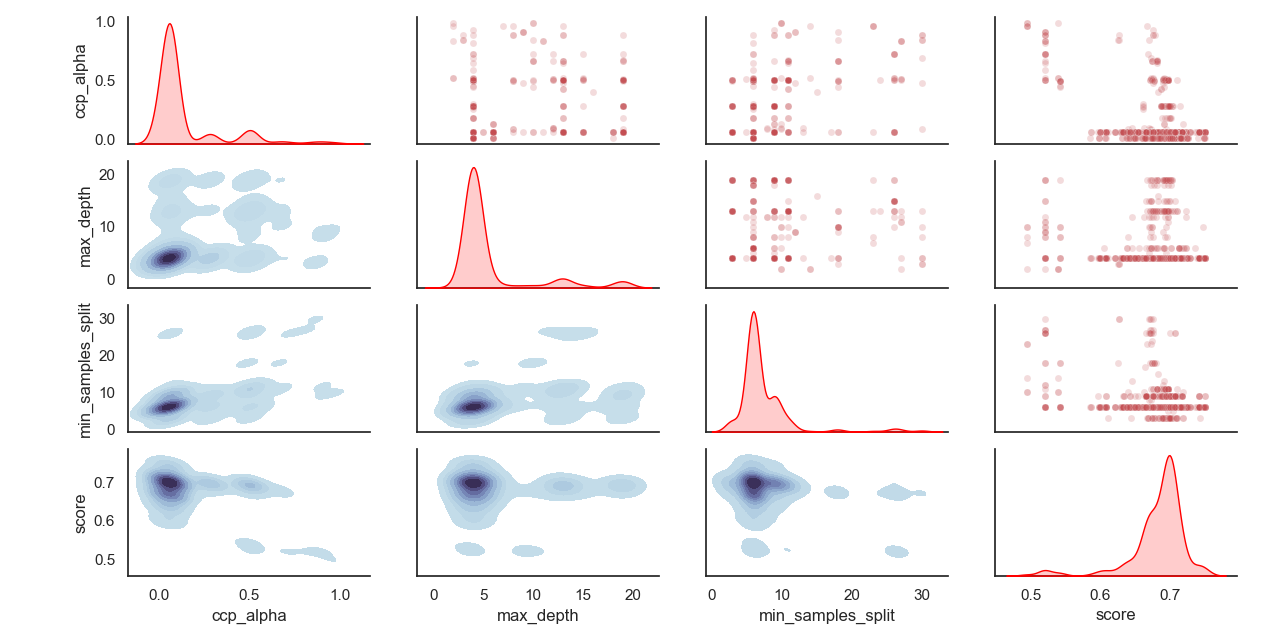
採樣超參數的分佈:
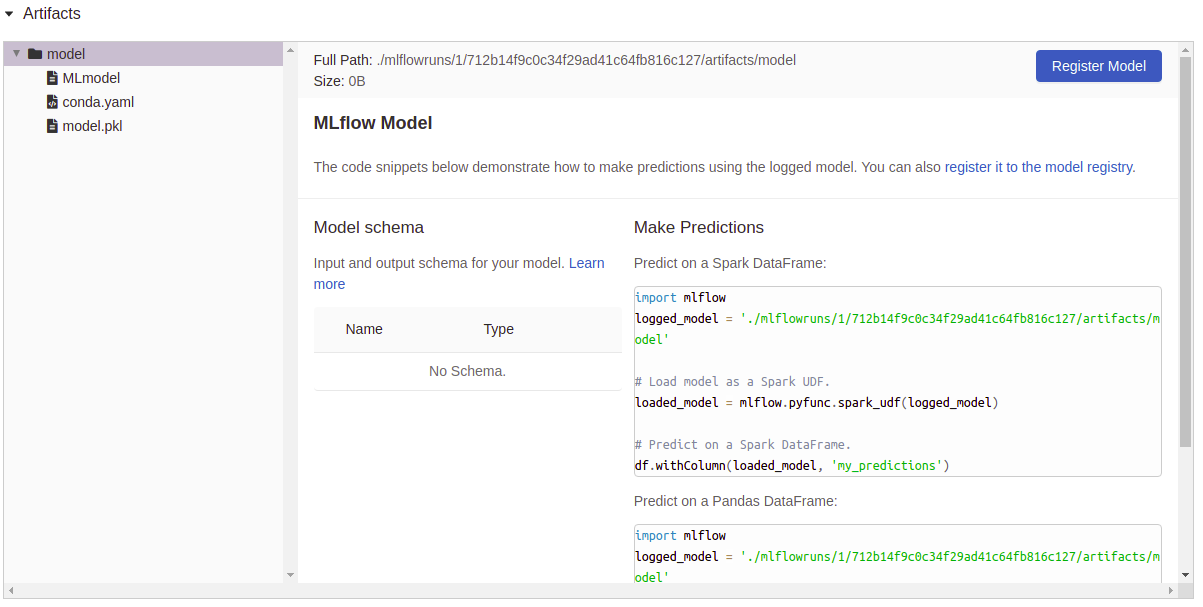
文物記錄:
安裝Sklearn-Genetic-Opt
建議在Env內使用虛擬Env安裝Sklearn-Genetic:
PIP安裝Sklearn-Genetic-Opt
如果您想獲得所有功能,包括繪圖,張板和MLFlow日誌記錄功能,請安裝所有額外的軟件包:
PIP安裝Sklearn-Genetic-Opt [全部]
from sklearn_genetic import GASearchCV
from sklearn_genetic . space import Continuous , Categorical , Integer
from sklearn . ensemble import RandomForestClassifier
from sklearn . model_selection import train_test_split , StratifiedKFold
from sklearn . datasets import load_digits
from sklearn . metrics import accuracy_score
data = load_digits ()
n_samples = len ( data . images )
X = data . images . reshape (( n_samples , - 1 ))
y = data [ 'target' ]
X_train , X_test , y_train , y_test = train_test_split ( X , y , test_size = 0.33 , random_state = 42 )
clf = RandomForestClassifier ()
# Defines the possible values to search
param_grid = { 'min_weight_fraction_leaf' : Continuous ( 0.01 , 0.5 , distribution = 'log-uniform' ),
'bootstrap' : Categorical ([ True , False ]),
'max_depth' : Integer ( 2 , 30 ),
'max_leaf_nodes' : Integer ( 2 , 35 ),
'n_estimators' : Integer ( 100 , 300 )}
# Seed solutions
warm_start_configs = [
{ "min_weight_fraction_leaf" : 0.02 , "bootstrap" : True , "max_depth" : None , "n_estimators" : 100 },
{ "min_weight_fraction_leaf" : 0.4 , "bootstrap" : True , "max_depth" : 5 , "n_estimators" : 200 },
]
cv = StratifiedKFold ( n_splits = 3 , shuffle = True )
evolved_estimator = GASearchCV ( estimator = clf ,
cv = cv ,
scoring = 'accuracy' ,
population_size = 20 ,
generations = 35 ,
param_grid = param_grid ,
n_jobs = - 1 ,
verbose = True ,
use_cache = True ,
warm_start_configs = warm_start_configs ,
keep_top_k = 4 )
# Train and optimize the estimator
evolved_estimator . fit ( X_train , y_train )
# Best parameters found
print ( evolved_estimator . best_params_ )
# Use the model fitted with the best parameters
y_predict_ga = evolved_estimator . predict ( X_test )
print ( accuracy_score ( y_test , y_predict_ga ))
# Saved metadata for further analysis
print ( "Stats achieved in each generation: " , evolved_estimator . history )
print ( "Best k solutions: " , evolved_estimator . hof ) from sklearn_genetic import GAFeatureSelectionCV , ExponentialAdapter
from sklearn . model_selection import train_test_split
from sklearn . svm import SVC
from sklearn . datasets import load_iris
from sklearn . metrics import accuracy_score
import numpy as np
data = load_iris ()
X , y = data [ "data" ], data [ "target" ]
# Add random non-important features
noise = np . random . uniform ( 5 , 10 , size = ( X . shape [ 0 ], 5 ))
X = np . hstack (( X , noise ))
X_train , X_test , y_train , y_test = train_test_split ( X , y , test_size = 0.33 , random_state = 0 )
clf = SVC ( gamma = 'auto' )
mutation_scheduler = ExponentialAdapter ( 0.8 , 0.2 , 0.01 )
crossover_scheduler = ExponentialAdapter ( 0.2 , 0.8 , 0.01 )
evolved_estimator = GAFeatureSelectionCV (
estimator = clf ,
scoring = "accuracy" ,
population_size = 30 ,
generations = 20 ,
mutation_probability = mutation_scheduler ,
crossover_probability = crossover_scheduler ,
n_jobs = - 1 )
# Train and select the features
evolved_estimator . fit ( X_train , y_train )
# Features selected by the algorithm
features = evolved_estimator . support_
print ( features )
# Predict only with the subset of selected features
y_predict_ga = evolved_estimator . predict ( X_test )
print ( accuracy_score ( y_test , y_predict_ga ))
# Transform the original data to the selected features
X_reduced = evolved_estimator . transform ( X_test )有關Sklearn-Genetic-Opt的變化的註釋,請參見Changelog
您可以使用命令檢查最新的開發版本:
git克隆https://github.com/rodrigo-arenas/sklearn-genetic-opt.git
安裝開發依賴性:
pip install -r dev -quirements.txt
檢查最新的開發文件:https://sklearn-genetic-opt.readthedocs.io/en/latest/
貢獻非常歡迎!正在進行的項目中有幾個機會,因此,如果您願意提供幫助,請聯繫。確保檢查當前問題以及貢獻指南。
非常感謝正在為這個項目提供幫助的人們!
安裝後,您可以從源目錄外啟動測試套件:
pytest sklearn_genetic