

Model scikit-learn Tuning hyperparameters dan pemilihan fitur, menggunakan algoritma evolusi.
Ini dimaksudkan untuk menjadi alternatif dari metode populer di dalam scikit-learn seperti pencarian grid dan pencarian grid acak untuk penyetelan hiperparameter, dan dari RFE (eliminasi fitur rekursif), pilih dari model untuk pemilihan fitur.
SKLEARN-genetic-opt menggunakan algoritma evolusioner dari paket DEAP (algoritma evolusioner terdistribusi dalam Python) untuk memilih himpunan hiperparameter yang mengoptimalkan (maks atau min) skor validasi silang, dapat digunakan untuk kedua masalah regresi dan klasifikasi.
Dokumentasi tersedia di sini
Visualisasikan kemajuan pelatihan Anda:
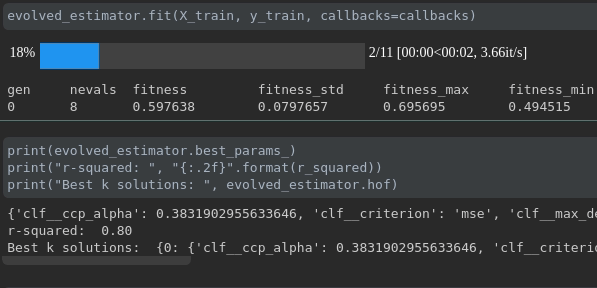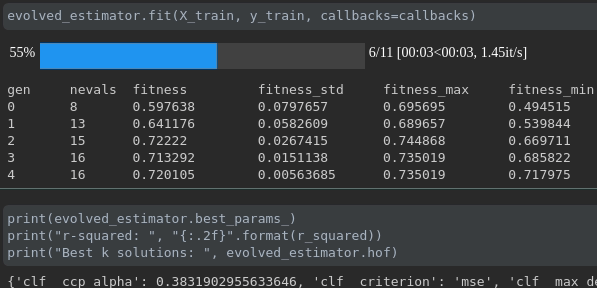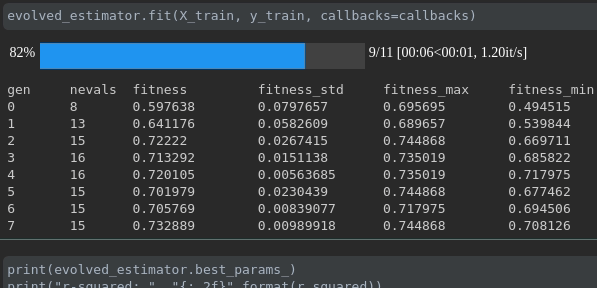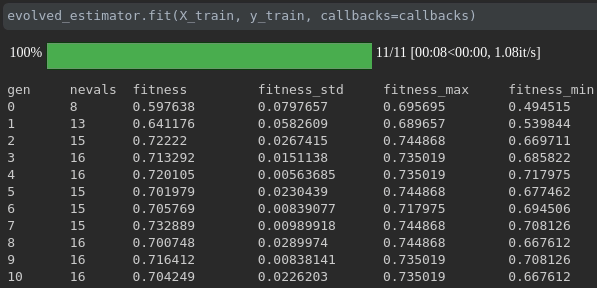
Visualisasi dan perbandingan metrik real-time lintas berjalan:
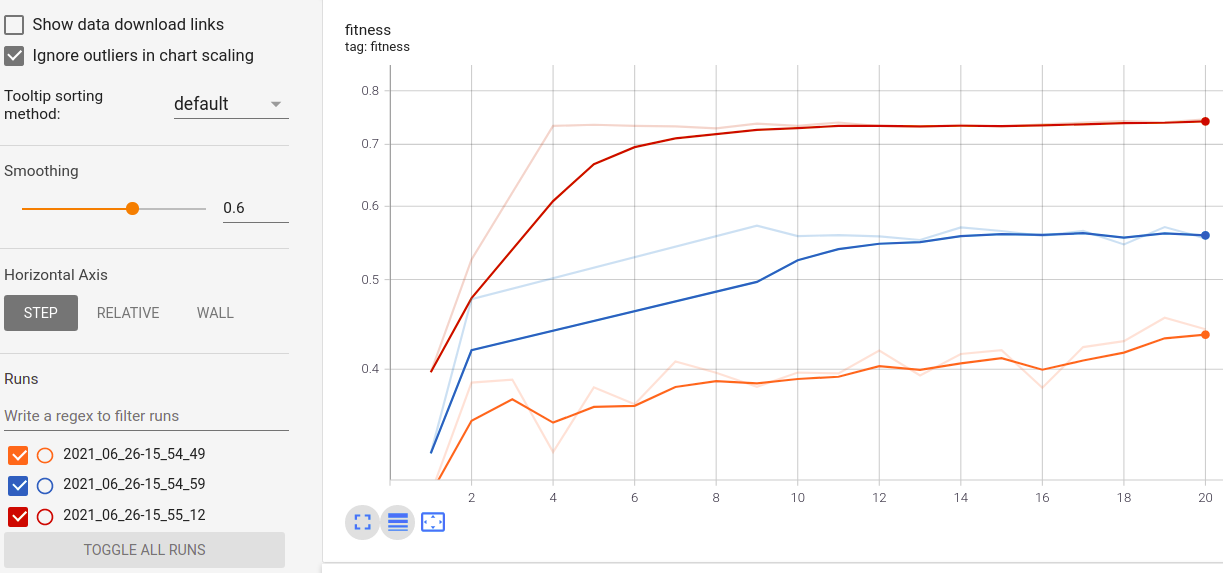
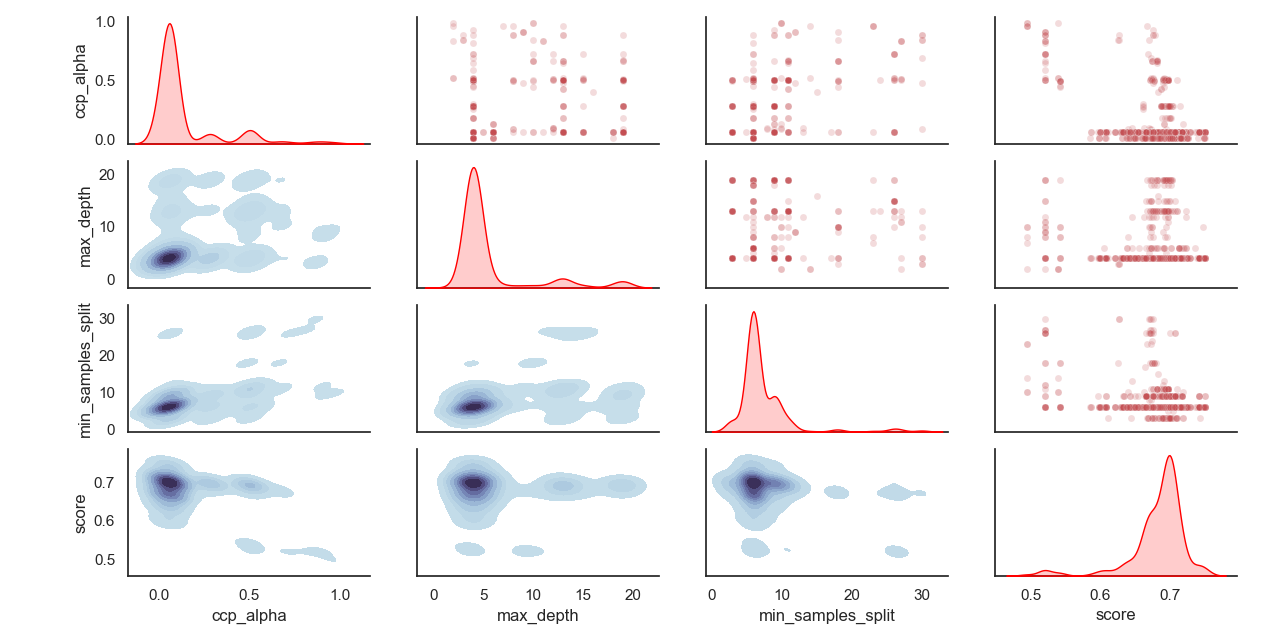
Distribusi HyperParameter yang Sampel:
Logging Artefak:
Instal sklearn-genetic-opt
Disarankan untuk menginstal sklearn-genetic menggunakan env virtual, di dalam env use:
PIP menginstal sklearn-genetic-opt
Jika Anda ingin mendapatkan semua fitur, termasuk plot, Tensorboard, dan kemampuan logging MLFLOW, pasang semua paket tambahan:
Pip menginstal sklearn-genetic-opt [semua]
from sklearn_genetic import GASearchCV
from sklearn_genetic . space import Continuous , Categorical , Integer
from sklearn . ensemble import RandomForestClassifier
from sklearn . model_selection import train_test_split , StratifiedKFold
from sklearn . datasets import load_digits
from sklearn . metrics import accuracy_score
data = load_digits ()
n_samples = len ( data . images )
X = data . images . reshape (( n_samples , - 1 ))
y = data [ 'target' ]
X_train , X_test , y_train , y_test = train_test_split ( X , y , test_size = 0.33 , random_state = 42 )
clf = RandomForestClassifier ()
# Defines the possible values to search
param_grid = { 'min_weight_fraction_leaf' : Continuous ( 0.01 , 0.5 , distribution = 'log-uniform' ),
'bootstrap' : Categorical ([ True , False ]),
'max_depth' : Integer ( 2 , 30 ),
'max_leaf_nodes' : Integer ( 2 , 35 ),
'n_estimators' : Integer ( 100 , 300 )}
# Seed solutions
warm_start_configs = [
{ "min_weight_fraction_leaf" : 0.02 , "bootstrap" : True , "max_depth" : None , "n_estimators" : 100 },
{ "min_weight_fraction_leaf" : 0.4 , "bootstrap" : True , "max_depth" : 5 , "n_estimators" : 200 },
]
cv = StratifiedKFold ( n_splits = 3 , shuffle = True )
evolved_estimator = GASearchCV ( estimator = clf ,
cv = cv ,
scoring = 'accuracy' ,
population_size = 20 ,
generations = 35 ,
param_grid = param_grid ,
n_jobs = - 1 ,
verbose = True ,
use_cache = True ,
warm_start_configs = warm_start_configs ,
keep_top_k = 4 )
# Train and optimize the estimator
evolved_estimator . fit ( X_train , y_train )
# Best parameters found
print ( evolved_estimator . best_params_ )
# Use the model fitted with the best parameters
y_predict_ga = evolved_estimator . predict ( X_test )
print ( accuracy_score ( y_test , y_predict_ga ))
# Saved metadata for further analysis
print ( "Stats achieved in each generation: " , evolved_estimator . history )
print ( "Best k solutions: " , evolved_estimator . hof ) from sklearn_genetic import GAFeatureSelectionCV , ExponentialAdapter
from sklearn . model_selection import train_test_split
from sklearn . svm import SVC
from sklearn . datasets import load_iris
from sklearn . metrics import accuracy_score
import numpy as np
data = load_iris ()
X , y = data [ "data" ], data [ "target" ]
# Add random non-important features
noise = np . random . uniform ( 5 , 10 , size = ( X . shape [ 0 ], 5 ))
X = np . hstack (( X , noise ))
X_train , X_test , y_train , y_test = train_test_split ( X , y , test_size = 0.33 , random_state = 0 )
clf = SVC ( gamma = 'auto' )
mutation_scheduler = ExponentialAdapter ( 0.8 , 0.2 , 0.01 )
crossover_scheduler = ExponentialAdapter ( 0.2 , 0.8 , 0.01 )
evolved_estimator = GAFeatureSelectionCV (
estimator = clf ,
scoring = "accuracy" ,
population_size = 30 ,
generations = 20 ,
mutation_probability = mutation_scheduler ,
crossover_probability = crossover_scheduler ,
n_jobs = - 1 )
# Train and select the features
evolved_estimator . fit ( X_train , y_train )
# Features selected by the algorithm
features = evolved_estimator . support_
print ( features )
# Predict only with the subset of selected features
y_predict_ga = evolved_estimator . predict ( X_test )
print ( accuracy_score ( y_test , y_predict_ga ))
# Transform the original data to the selected features
X_reduced = evolved_estimator . transform ( X_test )Lihat Changelog untuk Catatan tentang Perubahan SKLEARN-GENETIK-OPT
Anda dapat memeriksa versi pengembangan terbaru dengan perintah:
Git Clone https://github.com/rodrigo-arenas/sklearn-genetic-opt.git
Pasang dependensi pengembangan:
Pip instal -r dev -requirements.txt
Periksa dokumentasi in-development terbaru: https://sklearn-genetic-opt.readthedocs.io/en/latest/
Kontribusi lebih dari diterima! Ada beberapa peluang pada proyek yang sedang berlangsung, jadi silakan menghubungi jika Anda ingin membantu. Pastikan untuk memeriksa masalah saat ini dan juga panduan kontribusi.
Terima kasih banyak kepada orang -orang yang membantu proyek ini!
Setelah instalasi, Anda dapat meluncurkan test suite dari luar direktori sumber:
Pytest sklearn_genetic