存储库包含:
我们发布了DNABERT-S,这是一种基于DNABERT-2的基础模型,专门设计用于生成嵌入的DNA嵌入,该DNA自然簇和分离嵌入空间中不同物种的基因组。如果您有兴趣,请在此处查看。
DNABERT-2是一种基金会模型,该模型训练了大规模多种物种基因组,可实现最先进的性能
预先训练的模型可在Huggingface上使用zhihan1996/DNABERT-2-117M 。链接到HuggingFace ModelHub。链接直接下载。
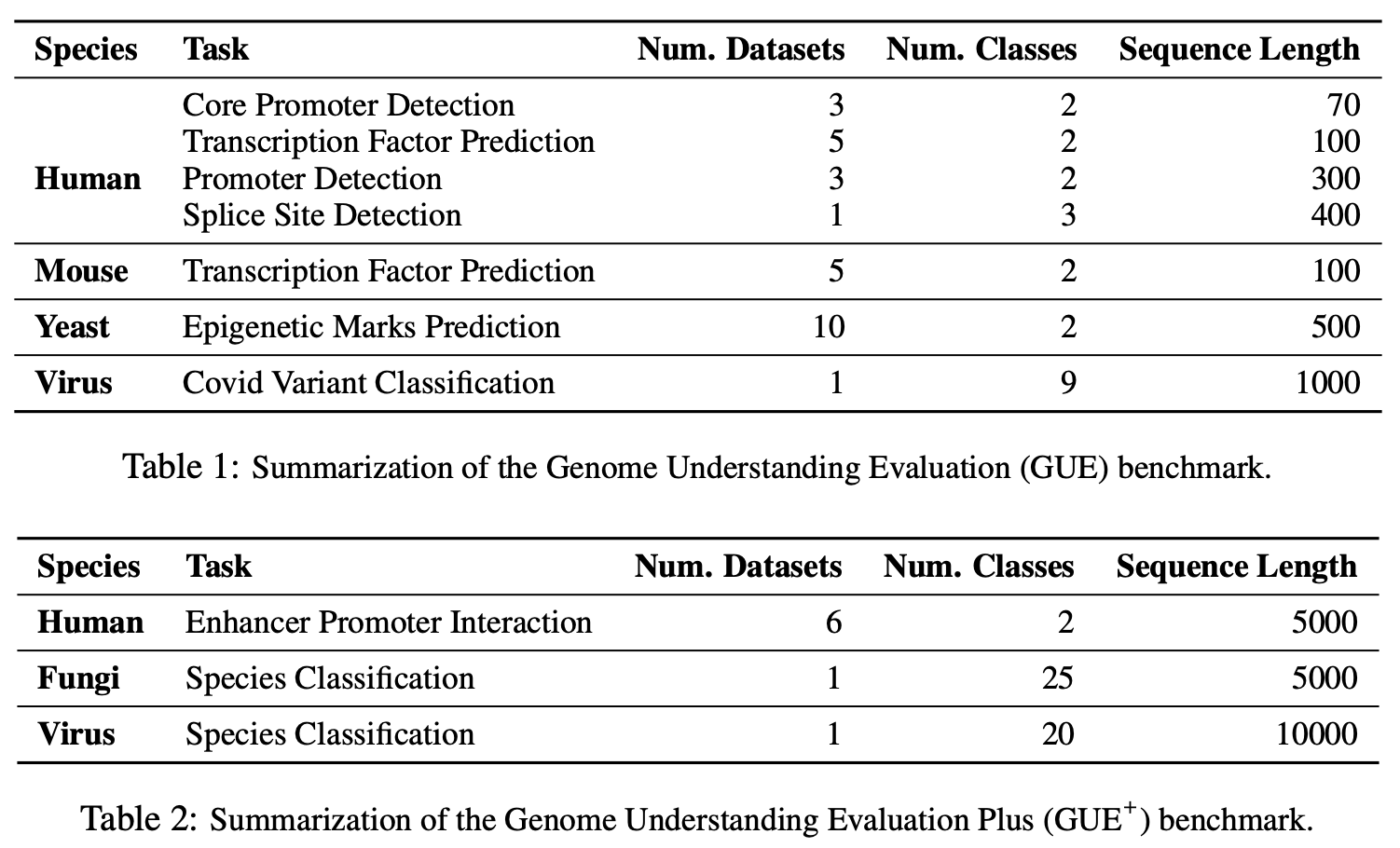
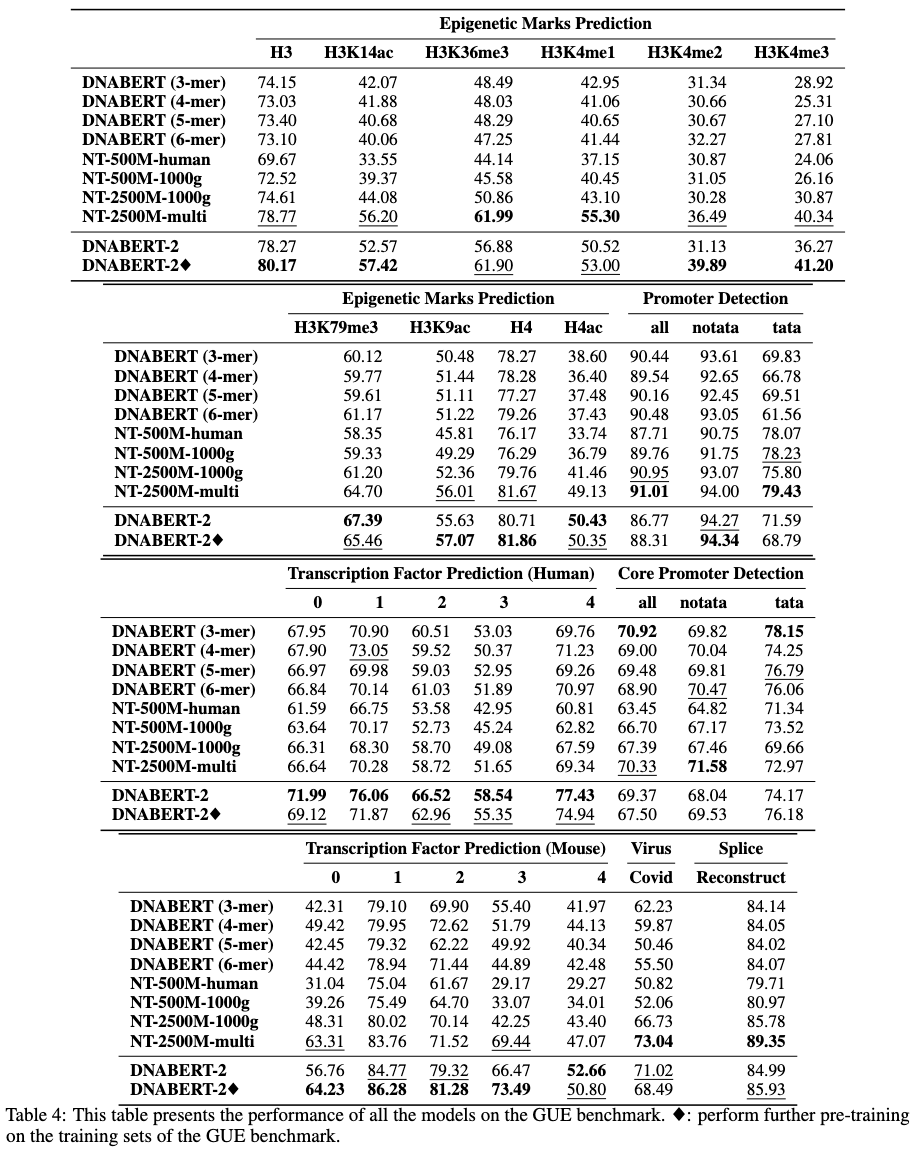
Gue是基因组理解的全面基准
# create and activate virtual python environment
conda create -n dna python=3.8
conda activate dna
# (optional if you would like to use flash attention)
# install triton from source
git clone https://github.com/openai/triton.git;
cd triton/python;
pip install cmake; # build-time dependency
pip install -e .
# install required packages
python3 -m pip install -r requirements.txt
我们的模型易于与Transformers软件包一起使用。
从HuggingFace(版本4.28)加载模型:
import torch
from transformers import AutoTokenizer , AutoModel
tokenizer = AutoTokenizer . from_pretrained ( "zhihan1996/DNABERT-2-117M" , trust_remote_code = True )
model = AutoModel . from_pretrained ( "zhihan1996/DNABERT-2-117M" , trust_remote_code = True )从HuggingFace(版本> 4.28)加载模型:
from transformers . models . bert . configuration_bert import BertConfig
config = BertConfig . from_pretrained ( "zhihan1996/DNABERT-2-117M" )
model = AutoModel . from_pretrained ( "zhihan1996/DNABERT-2-117M" , trust_remote_code = True , config = config )计算DNA序列的嵌入
dna = "ACGTAGCATCGGATCTATCTATCGACACTTGGTTATCGATCTACGAGCATCTCGTTAGC"
inputs = tokenizer(dna, return_tensors = 'pt')["input_ids"]
hidden_states = model(inputs)[0] # [1, sequence_length, 768]
# embedding with mean pooling
embedding_mean = torch.mean(hidden_states[0], dim=0)
print(embedding_mean.shape) # expect to be 768
# embedding with max pooling
embedding_max = torch.max(hidden_states[0], dim=0)[0]
print(embedding_max.shape) # expect to be 768
我们使用并稍微修改了Mosaicbert实施,用于DNABERT-2 https://github.com/mosaicml/examples/tree/main/main/main/examples/benchmarks/bert。您应该能够按照说明复制模型培训。
或者,您可以在https://github.com/huggingface/transformers/tree/main/main/examples/pytorch/language-modeling上使用Run_mlm.py它应该产生非常相似的模型。
培训数据可在此处获得。
请首先从此处下载GUE数据集。然后运行脚本以评估所有任务。
当前的脚本设置为使用DataParallel进行4个GPU进行培训。如果您有不同数量的GPU,请更改per_device_train_batch_size和gradient_accumulation_steps以相应地将全局批次大小调整为32,以复制论文中的结果。如果您想执行分布式的多GPU培训(例如,使用DistributedDataParallel ),只需将python更改为torchrun --nproc_per_node ${n_gpu}即可。
export DATA_PATH=/path/to/GUE #(e.g., /home/user)
cd finetune
# Evaluate DNABERT-2 on GUE
sh scripts/run_dnabert2.sh DATA_PATH
# Evaluate DNABERT (e.g., DNABERT with 3-mer) on GUE
# 3 for 3-mer, 4 for 4-mer, 5 for 5-mer, 6 for 6-mer
sh scripts/run_dnabert1.sh DATA_PATH 3
# Evaluate Nucleotide Transformers on GUE
# 0 for 500m-1000g, 1 for 500m-human-ref, 2 for 2.5b-1000g, 3 for 2.5b-multi-species
sh scripts/run_nt.sh DATA_PATH 0
在这里,我们提供了您自己数据集上微调DNABERT2的示例。
首先,请从数据集生成3个csv文件: train.csv , dev.csv和test.csv 。在培训过程中,该模型在train.csv上进行了培训,并在dev.csv文件上进行了评估。训练后,如果完成,则加载了dev.csv文件上最小损失的检查点,并在test.csv上进行评估。如果您没有验证集,请进行dev.csv和test.csv相同。
请参阅sample_data文件夹以获取数据格式示例。每个文件应采用相同的格式,第一行作为文档头名sequence, label 。以下每个行应包含由, (例如, ACGTCAGTCAGCGTACGT, 1 )串联的DNA序列和数值标记。
然后,您可以使用以下代码在您自己的数据集上进行Finetune dnabert-2:
cd finetune
export DATA_PATH=$path/to/data/folder # e.g., ./sample_data
export MAX_LENGTH=100 # Please set the number as 0.25 * your sequence length.
# e.g., set it as 250 if your DNA sequences have 1000 nucleotide bases
# This is because the tokenized will reduce the sequence length by about 5 times
export LR=3e-5
# Training use DataParallel
python train.py
--model_name_or_path zhihan1996/DNABERT-2-117M
--data_path ${DATA_PATH}
--kmer -1
--run_name DNABERT2_${DATA_PATH}
--model_max_length ${MAX_LENGTH}
--per_device_train_batch_size 8
--per_device_eval_batch_size 16
--gradient_accumulation_steps 1
--learning_rate ${LR}
--num_train_epochs 5
--fp16
--save_steps 200
--output_dir output/dnabert2
--evaluation_strategy steps
--eval_steps 200
--warmup_steps 50
--logging_steps 100
--overwrite_output_dir True
--log_level info
--find_unused_parameters False
# Training use DistributedDataParallel (more efficient)
export num_gpu=4 # please change the value based on your setup
torchrun --nproc_per_node=${num_gpu} train.py
--model_name_or_path zhihan1996/DNABERT-2-117M
--data_path ${DATA_PATH}
--kmer -1
--run_name DNABERT2_${DATA_PATH}
--model_max_length ${MAX_LENGTH}
--per_device_train_batch_size 8
--per_device_eval_batch_size 16
--gradient_accumulation_steps 1
--learning_rate ${LR}
--num_train_epochs 5
--fp16
--save_steps 200
--output_dir output/dnabert2
--evaluation_strategy steps
--eval_steps 200
--warmup_steps 50
--logging_steps 100
--overwrite_output_dir True
--log_level info
--find_unused_parameters False
如果您对我们的纸张或代码有任何疑问,请随时开始发行问题或发送电子邮件([email protected])。
如果您在工作中使用DNABERT-2,请请我们的论文:
DNABERT-2
@misc{zhou2023dnabert2,
title={DNABERT-2: Efficient Foundation Model and Benchmark For Multi-Species Genome},
author={Zhihan Zhou and Yanrong Ji and Weijian Li and Pratik Dutta and Ramana Davuluri and Han Liu},
year={2023},
eprint={2306.15006},
archivePrefix={arXiv},
primaryClass={q-bio.GN}
}
dnabert
@article{ji2021dnabert,
author = {Ji, Yanrong and Zhou, Zhihan and Liu, Han and Davuluri, Ramana V},
title = "{DNABERT: pre-trained Bidirectional Encoder Representations from Transformers model for DNA-language in genome}",
journal = {Bioinformatics},
volume = {37},
number = {15},
pages = {2112-2120},
year = {2021},
month = {02},
issn = {1367-4803},
doi = {10.1093/bioinformatics/btab083},
url = {https://doi.org/10.1093/bioinformatics/btab083},
eprint = {https://academic.oup.com/bioinformatics/article-pdf/37/15/2112/50578892/btab083.pdf},
}