该存储库包含以下论文的代码:
@misc{ziletti2024retrieval,
title={Retrieval augmented text-to-SQL generation for epidemiological question answering using electronic health records},
author={Angelo Ziletti and Leonardo D'Ambrosi},
year={2024},
eprint={2403.09226},
archivePrefix={arXiv},
primaryClass={cs.CL}
}
请在此处找到预印本:Ziletti和D'Ambrosi,https://arxiv.org/abs/2403.09226
本文在NAACL 2024临床NLP研讨会(https://clinical-nlp.github.io/2024/)上接受。
如果您在工作或研究中使用此代码,请引用这项工作。
这是该过程的摘要工作流程: 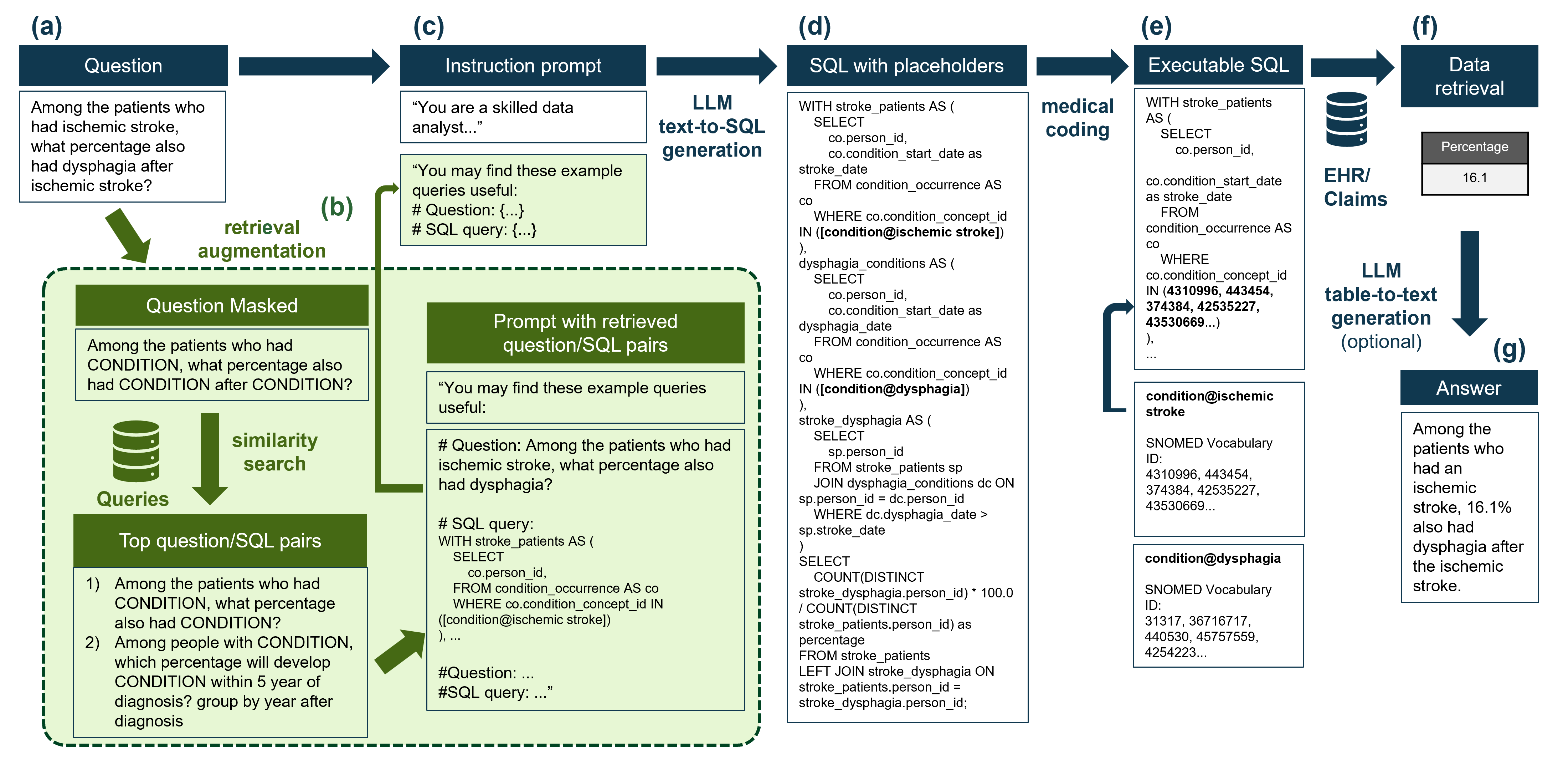 这是主要结果:
这是主要结果: 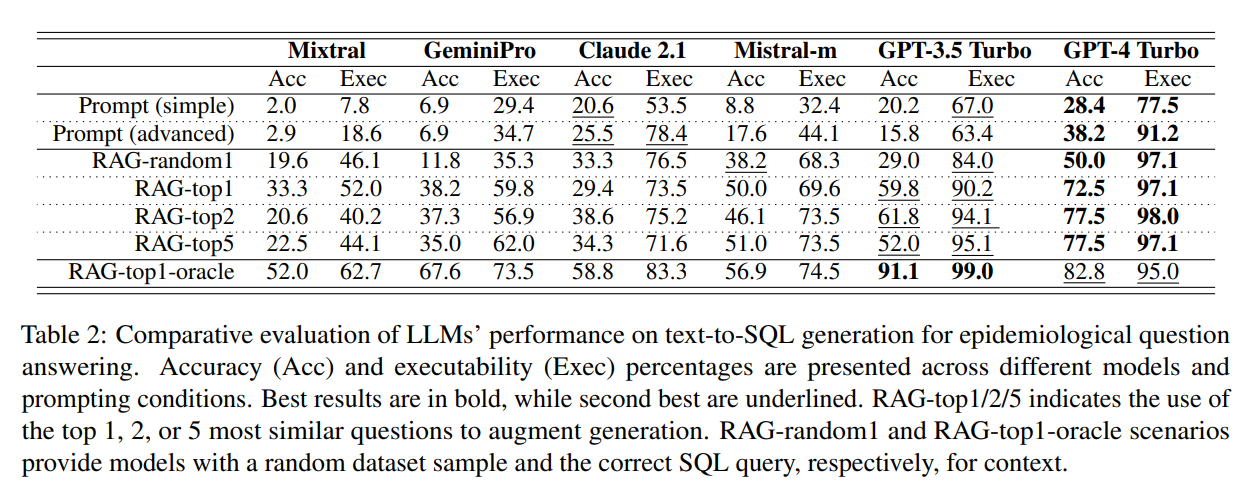
请参阅手稿以获取更多详细信息:https://arxiv.org/abs/2403.09226
标记的问题-SQL对在dataset集文件夹中的文件text2sql_epi_dataset_omop.xlsx中。
我们在实验中使用了Python 3.11。
从GitHub上的Master Branch安装最新版本:
git clone <GITHUB-URL>
cd text-to-sql-epi-ehr-naacl2024
pip install -r requirements.txt
首先,我们需要从dataset文件夹中提供的Excel文件创建一个腌制文件(查询库)
cd scripts
python run_querylib_calc.py
然后将使用此泡菜文件在查询生成阶段执行检索增强生成(RAG)。
要执行预测,请用双引号以您的问题运行脚本prediction_pipeline.py 。例如,
cd scripts
python prediction_pipeline.py --question "How many women with atopic dermatitis?"
这应该返回:
Question: How many women with atopic dermatitis?
SQL template:
SELECT COUNT(DISTINCT p.person_id) AS female_patients_with_atopic_dermatitis
FROM condition_occurrence AS co
JOIN person AS p ON co.person_id = p.person_id
WHERE co.condition_concept_id IN ([condition@atopic dermatitis])
AND p.gender_concept_id = 8532;
要执行查询,您将是Google Cloud上的数据集的链接:https://console.cloud.google.com/marketplace/product/product/hhs/synpuf您可以打开链接(需要Google帐户),并在Google帐户中运行查询。
要使查询可在Google BigQuery上执行,您将需要修改表的生成名称以符合Google查询表名称。这只是桌子的重命名。
例如,
SELECT COUNT(DISTINCT p.person_id) AS female_patients_with_atopic_dermatitis
FROM person AS p
JOIN condition_occurrence AS co ON p.person_id = co.person_id
JOIN concept AS c ON co.condition_concept_id = c.concept_id
WHERE p.gender_concept_id = 8532
AND c.concept_id IN (133834,4298597,4066382,4298599,4296193,4080929,4296192,4290738,4290734,4206125,4290736,4080928,4033671,4031630,4297478,4296190,4031631,4080927,4298598,4298601,4031013,4297362,4290740,4297495,40482226,4298600,4236759);
必须更改为
SELECT
COUNT(DISTINCT p.person_id) AS female_patients_with_atopic_dermatitis
FROM `bigquery-public-data.cms_synthetic_patient_data_omop.person` AS p
JOIN `bigquery-public-data.cms_synthetic_patient_data_omop.condition_occurrence` AS co ON p.person_id = co.person_id
JOIN `bigquery-public-data.cms_synthetic_patient_data_omop.concept` AS c ON co.condition_concept_id = c.concept_id
WHERE p.gender_concept_id = 8532
AND c.concept_id IN (133834,4298597,4066382,4298599,4296193,4080929,4296192,4290738,4290734,4206125,4290736,4080928,4033671,4031630,4297478,4296190,4031631,4080927,4298598,4298601,4031013,4297362,4290740,4297495,40482226,4298600,4236759);
在上面的BigQuery实例上运行。
如果您有自己的雪花数据仓库,则可能不需要执行这些更改。
在存储库中,我们为医学编码提供了一个模型版本,以显示如何将医疗编码整合到过程中,如上图所示。编码是在SNOMED本体中执行的,因为这是存储在OMOP公共数据模型(OMOP-CDM)中的数据的基本本体。该表通常在OMOP-CDM中称为Concept_table。
我们在dataset集文件夹中准备了一个称为medcodes_mockup.xlsx的小型模型表。在这里,我们根据OMOP-CDM的数据结构提供合成数据。您可以在此处从国家医学图书馆下载整个本体。 (请注意,您需要接受他们的条款和条件)
例如,可以在此处找到有关Snomed-CT的更多信息
首先,我们需要从提供的Excel文件中创建一个腌制文件(Medcodeonto库)
cd scripts
python run_medcoding_calc.py
然后将使用此腌制文件来搜索腌制文件中的医疗代码。运行文件还将提供一个示例,说明如何执行编码。
请注意,这只是模型实现。对于准备生产的版本,我们建议使用矢量数据库(例如QDRANT),例如从雪花数据库中为整个概念表索引。
运行Medical coding compilation步骤后,您可以运行完整的管道:SQL查询生成 +医疗编码。
这是一个例子:
cd scripts
python prediction_pipeline.py --med_coding True --question "How many females with atopic dermatitis"
这将返回
Use medical coding: True
Use Snowflake database: False
No sentence-transformers model found with name BAAI/bge-large-en-v1.5. Creating a new one with MEAN pooling.
Question: How many females with atopic dermatitis
SQL template:
SELECT COUNT(DISTINCT p.person_id) AS female_patients_with_atopic_dermatitis
FROM condition_occurrence AS co
JOIN person AS p ON co.person_id = p.person_id
WHERE co.condition_concept_id IN ([condition@atopic dermatitis])
AND p.gender_concept_id = 8532;
Loading embedding from C:UsersGKENYOneDrive - BayerPersonal Dataascenttext-to-sql-epi-ehr-naacl2024data_outmedcodes_onto.pkl
Retrieved codes: {'atopic dermatitis': [{'Score': 0.8221014142036438, 'CONCEPT_NAME': 'Dermatitis in children', 'CONCEPT_ID': 4296192, 'DOMAIN_ID': 'Condition', 'VOCABULARY_ID': 'SNOMED', 'STANDARD_CONCEPT': 'S', 'CONCEPT_CODE': 402196005}, {'Score': 0.8056577444076538, 'CONCEPT_NAME': 'Widespread dermatitis', 'CONCEPT_ID': 4298597, 'DOMAIN_ID': 'Condition', 'VOCABULARY_ID': 'SNOMED', 'STANDARD_CONCEPT': 'S', 'CONCEPT_CODE': 402186001}, {'Score': 0.7999188899993896, 'CONCEPT_NAME': 'Early childhood dermatitis', 'CONCEPT_ID': 4298599, 'DOMAIN_ID': 'Condition', 'VOCABULARY_ID': 'SNOMED', 'STANDARD_CONCEPT': 'S', 'CONCEPT_CODE': 402195009}, {'Score': 0.7992758750915527, 'CONCEPT_NAME': 'Facial dermatitis', 'CONCEPT_ID': 4298598, 'DOMAIN_ID': 'Condition', 'VOCABULARY_ID': 'SNOMED', 'STANDARD_CONCEPT': 'S', 'CONCEPT_CODE': 402192007}]}
Please note that the medical coding is based on a mockup ontology. Results will not be reliable
SQL filled:
SELECT COUNT(DISTINCT p.person_id) AS female_patients_with_atopic_dermatitis
FROM condition_occurrence AS co
JOIN person AS p ON co.person_id = p.person_id
WHERE co.condition_concept_id IN (4296192,4298597,4298599,4298598)
AND p.gender_concept_id = 8532;
请注意,医疗编码步骤仅在这里示例性。为了获得可靠的结果,您需要访问完整的SNOMED本体(请参阅Medical coding compilation部分)。