Periplus is currently in alpha and is not production-ready. The project is under active development, and is not yet recommended for use in production systems.
Periplus is an open-source in-memory vector database cache built on Meta's vector similarity search library FAISS. The project can best be thought of as "Redis for vector databases". It's designed to store a dynamically updated subset of a large vector collection entirely in memory while serving queries without interacting with any other nodes at query time. When Periplus receives a query, it first assesses whether it has the relevant part of the index in-residence. If it does, it resolves the query with the appropriate response. If it doesn't, it returns a cache miss and leaves the querier to fetch the data from the database. Periplus is not designed to function in isolation. Instead, it is meant to form a modular and flexible caching layer for a separate vector database which forms the persistance layer. The purpose of this is to enable lower latency and easy horizontal scaling for increasing throughput. For a more detailed description of the inspiration behind Periplus and how it works you can read the announcement blog: Introducing Periplus: A New Approach to Vector Database Caching.
Periplus uses an inverted file index (IVF) as the basis for cache management. Inverted file indexes partition the vector space into contiguous cells defined by a set of centroid vectors where each cell is defined as the region which is closer to it's centroid than to any other centroid. Queries are then resolved by first computing the distances from the query vector to the set of centroids and then searching only the cells defined by the n_probe (search hyperparameter) closest centroids. Periplus takes advantage of this by keeping a subset of these cells in residence at any given time and only resolving queries which are relevant to that subset while rejecting the ones that aren't as cache misses. Periplus loads and evicts entire IVF cells at a time to maintain the integrity of the index and ensure equivalent recall (on cache hits) to a standard IVF index. IVF cells are loaded by querying the vector database via a proxy with a list of ids of vectors which Periplus maintains to track which vectors occupy which cells. These operations can be invoked by the user using LOAD, SEARCH, and EVICT commands. For details, see the Periplus Commands section below.
Periplus can either be run as a Docker container or it can be built from source and run as an executable. No official binaries are currently available. Running Periplus as a container is the recommended approach, but both are viable options.
Currently, the Docker image only supports AMD64 architectures. This constraint stems from the base image, but more architectures will be supported in the near future. There are 2 ways to run Periplus as a container: download the official Docker image from Dockerhub (recommended) or build the image yourself. The first step in either case is to install Docker if you haven't already. The instructions to do so can be found here.
docker image pull qdl123/periplus:latest.docker run -p 3000:3000 qdl123/periplus:latestgit clone https://github.com/QDL123/Periplus.gitcd <path-to-periplus-repo>/Periplusdocker build -t periplus-image .docker run -p 3000:3000 periplus-image .Periplus uses CMake for it's build system. It expects all dependencies to have pre-compiled binaries installed via Homebrew. Homebrew is supported by MacOS, Ubuntu, and WSL if you're on Windows. Periplus has been built on MacOS/ARM64 and Ubuntu/AMD64. All other operating system and architecture combinations are untested. To build Periplus from source, follow the following steps:
brew install faiss curl cpr rapidjson libomp catch2 cmakegit clone https://github.com/QDL123/Periplus.gitcd <path-to-periplus-repo>/Peripluscmake -S . -B buildcmake --build build./build/periplus -p 3000Any system using Periplus will consist of 4 components: the vector database, a database proxy which allows Periplus to load data from the database, a Periplus instance, and a client application.
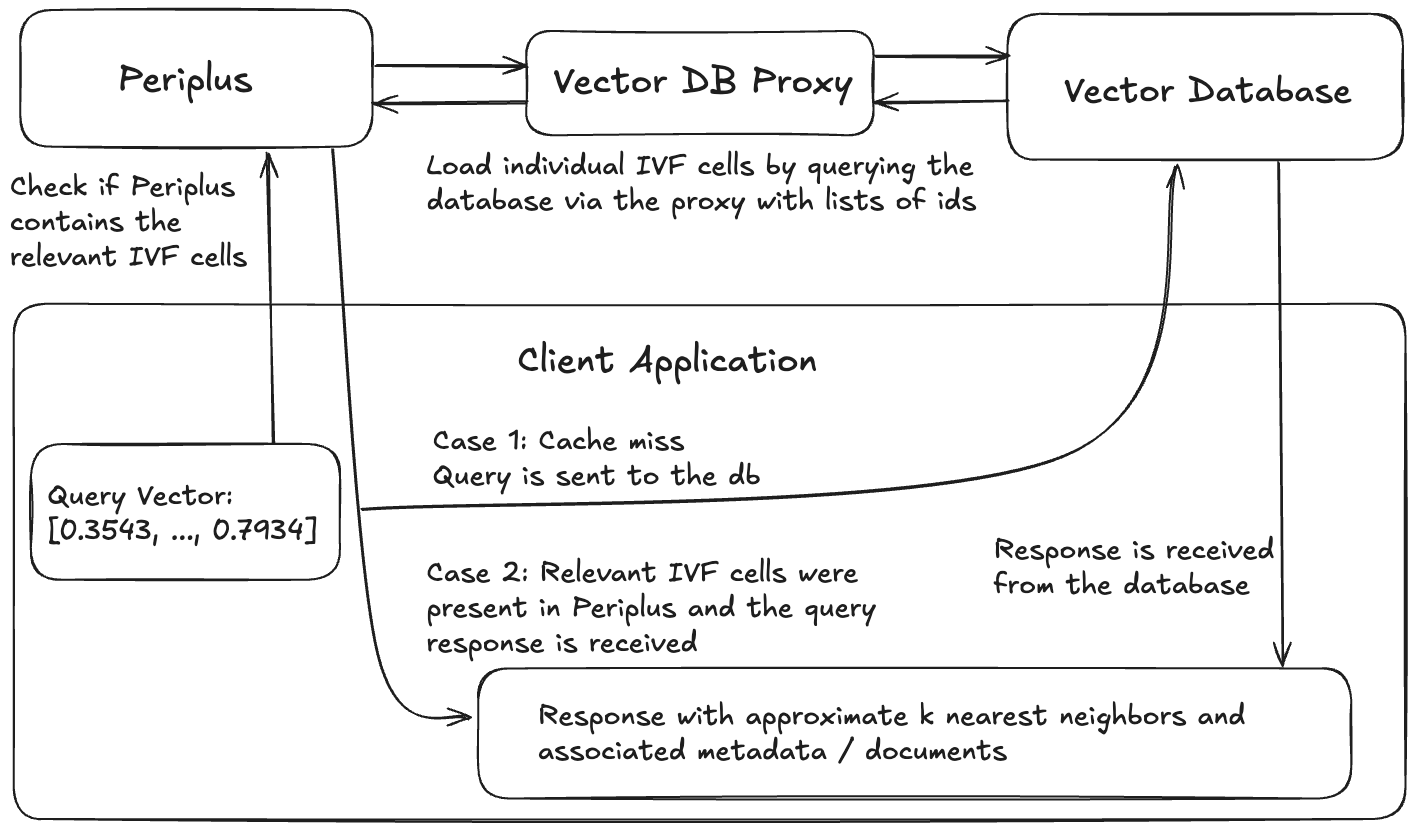
Example architecture of an application using Periplus.
Any vector database that allows for looking up data by a unique identifier (virtually all of them) will work. Periplus is designed to be most beneficial when working with really large vector collectons (billion-scale) where the index has to live on the file system as opposed to RAM, although that's not a requirement.
The vector database proxy's purpose is to provide a consistent interface for Periplus to interact with the vector database through. The proxy must implement a REST interface which accepts POST requests of the following form:
URL: This is flexible and can be specified by the Periplus client.
Headers:
"Content-Type": "application/json
Body:
{
"ids": ["id-1", "id-2", "id-3"]
}Response:
{
"results": [
{
"id": "String",
"embedding": [0.1, 0.2, 0.3],
"document": "String",
"metdata": "String"
}
]
}To make implementing this endpoint easier, you can you use the periplus-proxy python package which uses FastAPI to set everything up. All the user has to do is implement the following function and pass it as an argument:
async def fetch_ids(request: Query) -> QueryResult
For details on how to do this, you can check out the periplus-proxy package README.md.
Follow the instructions above to start up a Periplus instance.
To interact with your Periplus instance, use the Periplus client library. Currently only python is supported. For details on the client library, you can view it's README.md.
from periplus_client import Periplus
# host, port
client = Periplus("localhost", 13)
# vector dimensionality, database proxy url, options: (nTotal)
await client.initialize(d=d, db_url=url, options={ "nTotal": 50000 })
training_data = [[0.43456, ..., 0.38759], ...]
await client.train(training_data)
ids = ["0", ..., "n"]
embeddings = [[0.43456, ..., 0.38759], ...]
await client.add(ids=ids, embeddings=embeddings)
load_options = { "n_load": 2 }
# query_vector, optional: options object
await client.load([embeddings[0]] load_options)
# k, query_vector
response = await client.search(5, [embeddings[0]])
print(response)
'''
[ // Results for each of the n query vectors in xq
[ // K nearest neighbors to this corresponding index in the xq list
(
id="n",
embedding=[0.43456, ..., 0.38759],
document="",
metadata="{}"
),
...
],
...
]
'''
# query_vector
await client.evict(embeddings[0])We welcome contributions to Periplus! To learn how to get started, take a look at the Contribution Guide.
This project is licensed under the MIT License - see the LICENSE file for details.