现在在线纸! https://arxiv.org/abs/2111.00595
现在在线文档! https://mlmed.org/torchxrayvision/
| (?促销视频) ) |
|---|
用于胸部X射线数据集和型号的库。包括预训练的模型。
TorchxRayVision是一个开源软件库,用于使用胸部X射线数据集和深度学习模型。它为一组广泛的公共胸部X射线数据集提供了一个常见的接口和常见的预处理链。此外,可以通过库作为基准或特征提取器提供许多具有不同架构的分类和具有不同体系结构的表示模型。
Twitter:@torchxrayvision
$ pip install torchxrayvision
import torchxrayvision as xrv
import skimage , torch , torchvision
# Prepare the image:
img = skimage . io . imread ( "16747_3_1.jpg" )
img = xrv . datasets . normalize ( img , 255 ) # convert 8-bit image to [-1024, 1024] range
img = img . mean ( 2 )[ None , ...] # Make single color channel
transform = torchvision . transforms . Compose ([ xrv . datasets . XRayCenterCrop (), xrv . datasets . XRayResizer ( 224 )])
img = transform ( img )
img = torch . from_numpy ( img )
# Load model and process image
model = xrv . models . DenseNet ( weights = "densenet121-res224-all" )
outputs = model ( img [ None ,...]) # or model.features(img[None,...])
# Print results
dict ( zip ( model . pathologies , outputs [ 0 ]. detach (). numpy ()))
{ 'Atelectasis' : 0.32797316 ,
'Consolidation' : 0.42933336 ,
'Infiltration' : 0.5316924 ,
'Pneumothorax' : 0.28849724 ,
'Edema' : 0.024142697 ,
'Emphysema' : 0.5011832 ,
'Fibrosis' : 0.51887786 ,
'Effusion' : 0.27805611 ,
'Pneumonia' : 0.18569896 ,
'Pleural_Thickening' : 0.24489835 ,
'Cardiomegaly' : 0.3645515 ,
'Nodule' : 0.68982 ,
'Mass' : 0.6392845 ,
'Hernia' : 0.00993878 ,
'Lung Lesion' : 0.011150705 ,
'Fracture' : 0.51916164 ,
'Lung Opacity' : 0.59073937 ,
'Enlarged Cardiomediastinum' : 0.27218717 }示例脚本以处理图像使用概述的模型是process_image.py
$ python3 process_image.py ../tests/00000001_000.png
{'preds': {'Atelectasis': 0.50500506,
'Cardiomegaly': 0.6600903,
'Consolidation': 0.30575264,
'Edema': 0.274184,
'Effusion': 0.4026162,
'Emphysema': 0.5036339,
'Enlarged Cardiomediastinum': 0.40989172,
'Fibrosis': 0.53293407,
'Fracture': 0.32376793,
'Hernia': 0.011924741,
'Infiltration': 0.5154413,
'Lung Lesion': 0.22231922,
'Lung Opacity': 0.2772148,
'Mass': 0.32237658,
'Nodule': 0.5091847,
'Pleural_Thickening': 0.5102617,
'Pneumonia': 0.30947986,
'Pneumothorax': 0.24847917}}
指定预验证模型的权重(当前全部densenet121)注意:每个验证的模型都有18个输出。 all模型都有训练的每个输出。但是,对于其他权重,某些目标没有受过训练,并且会预测它们在训练数据集中不存在。唯一有效的输出在{dataset}.pathologies 。
## 224x224 models
model = xrv . models . DenseNet ( weights = "densenet121-res224-all" )
model = xrv . models . DenseNet ( weights = "densenet121-res224-rsna" ) # RSNA Pneumonia Challenge
model = xrv . models . DenseNet ( weights = "densenet121-res224-nih" ) # NIH chest X-ray8
model = xrv . models . DenseNet ( weights = "densenet121-res224-pc" ) # PadChest (University of Alicante)
model = xrv . models . DenseNet ( weights = "densenet121-res224-chex" ) # CheXpert (Stanford)
model = xrv . models . DenseNet ( weights = "densenet121-res224-mimic_nb" ) # MIMIC-CXR (MIT)
model = xrv . models . DenseNet ( weights = "densenet121-res224-mimic_ch" ) # MIMIC-CXR (MIT)
# 512x512 models
model = xrv . models . ResNet ( weights = "resnet50-res512-all" )
# DenseNet121 from JF Healthcare for the CheXpert competition
model = xrv . baseline_models . jfhealthcare . DenseNet ()
# Official Stanford CheXpert model
model = xrv . baseline_models . chexpert . DenseNet ( weights_zip = "chexpert_weights.zip" )
# Emory HITI lab race prediction model
model = xrv . baseline_models . emory_hiti . RaceModel ()
model . targets - > [ "Asian" , "Black" , "White" ]
# Riken age prediction model
model = xrv . baseline_models . riken . AgeModel ()模式的基准在这里:Benchmarks.md和某些模型的性能可以在本文arxiv.org/abs/2002.02497中看到。
您还可以加载预先训练的自动编码器,该自动编码器在Padchest,NIH,Chexpert和Mimic数据集上进行了训练。
ae = xrv . autoencoders . ResNetAE ( weights = "101-elastic" )
z = ae . encode ( image )
image2 = ae . decode ( z )您可以加载预贴的解剖分割模型。演示笔记本
seg_model = xrv . baseline_models . chestx_det . PSPNet ()
output = seg_model ( image )
output . shape # [1, 14, 512, 512]
seg_model . targets # ['Left Clavicle', 'Right Clavicle', 'Left Scapula', 'Right Scapula',
# 'Left Lung', 'Right Lung', 'Left Hilus Pulmonis', 'Right Hilus Pulmonis',
# 'Heart', 'Aorta', 'Facies Diaphragmatica', 'Mediastinum', 'Weasand', 'Spine'] 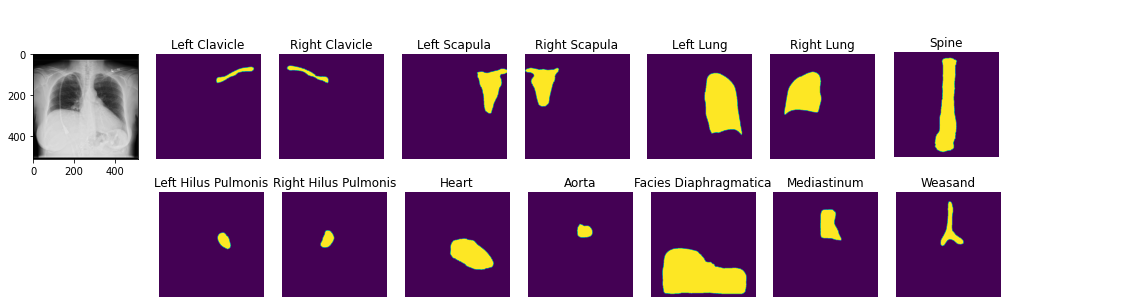
在每个数据集和演示笔记本上查看docstrings,以获取更多详细信息,并示例加载脚本
transform = torchvision . transforms . Compose ([ xrv . datasets . XRayCenterCrop (),
xrv . datasets . XRayResizer ( 224 )])
# RSNA Pneumonia Detection Challenge. https://pubs.rsna.org/doi/full/10.1148/ryai.2019180041
d_kaggle = xrv . datasets . RSNA_Pneumonia_Dataset ( imgpath = "path to stage_2_train_images_jpg" ,
transform = transform )
# CheXpert: A Large Chest Radiograph Dataset with Uncertainty Labels and Expert Comparison. https://arxiv.org/abs/1901.07031
d_chex = xrv . datasets . CheX_Dataset ( imgpath = "path to CheXpert-v1.0-small" ,
csvpath = "path to CheXpert-v1.0-small/train.csv" ,
transform = transform )
# National Institutes of Health ChestX-ray8 dataset. https://arxiv.org/abs/1705.02315
d_nih = xrv . datasets . NIH_Dataset ( imgpath = "path to NIH images" )
# A relabelling of a subset of NIH images from: https://pubs.rsna.org/doi/10.1148/radiol.2019191293
d_nih2 = xrv . datasets . NIH_Google_Dataset ( imgpath = "path to NIH images" )
# PadChest: A large chest x-ray image dataset with multi-label annotated reports. https://arxiv.org/abs/1901.07441
d_pc = xrv . datasets . PC_Dataset ( imgpath = "path to image folder" )
# COVID-19 Image Data Collection. https://arxiv.org/abs/2006.11988
d_covid19 = xrv . datasets . COVID19_Dataset () # specify imgpath and csvpath for the dataset
# SIIM Pneumothorax Dataset. https://www.kaggle.com/c/siim-acr-pneumothorax-segmentation
d_siim = xrv . datasets . SIIM_Pneumothorax_Dataset ( imgpath = "dicom-images-train/" ,
csvpath = "train-rle.csv" )
# VinDr-CXR: An open dataset of chest X-rays with radiologist's annotations. https://arxiv.org/abs/2012.15029
d_vin = xrv . datasets . VinBrain_Dataset ( imgpath = ".../train" ,
csvpath = ".../train.csv" )
# National Library of Medicine Tuberculosis Datasets. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4256233/
d_nlmtb = xrv . datasets . NLMTB_Dataset ( imgpath = "path to MontgomerySet or ChinaSet_AllFiles" )每个数据集都包含许多字段。使用xrv.datasets.subset_dataset和xrv.datasets.merge_dataset时,将保持这些字段。
.pathologies .labels
.labels该字段包含1,0或NAN .pathologies NAN。
.csv此字段是数据随附的元数据CSV文件的熊猫数据框架。每行都与数据集的元素对齐,因此使用.iloc索引将起作用。
如果可能的话,每个数据集的.csv将具有CSV的一些常见字段。当列表如下时,这些将被对齐:
csv.patientid一个独特的ID,该ID将在此数据集中单一识别样本
csv.offset_day_int在几天内图像的整数偏移。这预计将是相对时间,并且没有绝对含义,尽管对于某些数据集,这是时期的时间。
csv.age_years将患者的年龄多年。
csv.sex_male如果患者是男性
csv.sex_female如果患者是女性
relabel_dataset将使标签具有与病理参数相同的顺序。
xrv . datasets . relabel_dataset ( xrv . datasets . default_pathologies , d_nih ) # has side effects指定视图子集(演示笔记本)
d_kaggle = xrv . datasets . RSNA_Pneumonia_Dataset ( imgpath = "..." ,
views = [ "PA" , "AP" , "AP Supine" ])每个患者仅指定1张图像
d_kaggle = xrv . datasets . RSNA_Pneumonia_Dataset ( imgpath = "..." ,
unique_patients = True )每个数据集获取摘要统计信息
d_chex = xrv . datasets . CheX_Dataset ( imgpath = "CheXpert-v1.0-small" ,
csvpath = "CheXpert-v1.0-small/train.csv" ,
views = [ "PA" , "AP" ], unique_patients = False )
CheX_Dataset num_samples = 191010 views = [ 'PA' , 'AP' ]
{ 'Atelectasis' : { 0.0 : 17621 , 1.0 : 29718 },
'Cardiomegaly' : { 0.0 : 22645 , 1.0 : 23384 },
'Consolidation' : { 0.0 : 30463 , 1.0 : 12982 },
'Edema' : { 0.0 : 29449 , 1.0 : 49674 },
'Effusion' : { 0.0 : 34376 , 1.0 : 76894 },
'Enlarged Cardiomediastinum' : { 0.0 : 26527 , 1.0 : 9186 },
'Fracture' : { 0.0 : 18111 , 1.0 : 7434 },
'Lung Lesion' : { 0.0 : 17523 , 1.0 : 7040 },
'Lung Opacity' : { 0.0 : 20165 , 1.0 : 94207 },
'Pleural Other' : { 0.0 : 17166 , 1.0 : 2503 },
'Pneumonia' : { 0.0 : 18105 , 1.0 : 4674 },
'Pneumothorax' : { 0.0 : 54165 , 1.0 : 17693 },
'Support Devices' : { 0.0 : 21757 , 1.0 : 99747 }}以下数据集可用面具:
xrv . datasets . RSNA_Pneumonia_Dataset () # for Lung Opacity
xrv . datasets . SIIM_Pneumothorax_Dataset () # for Pneumothorax
xrv . datasets . NIH_Dataset () # for Cardiomegaly, Mass, Effusion, ...示例用法:
d_rsna = xrv . datasets . RSNA_Pneumonia_Dataset ( imgpath = "stage_2_train_images_jpg" ,
views = [ "PA" , "AP" ],
pathology_masks = True )
# The has_masks column will let you know if any masks exist for that sample
d_rsna . csv . has_masks . value_counts ()
False 20672
True 6012
# Each sample will have a pathology_masks dictionary where the index
# of each pathology will correspond to a mask of that pathology (if it exists).
# There may be more than one mask per sample. But only one per pathology.
sample [ "pathology_masks" ][ d_rsna . pathologies . index ( "Lung Opacity" )]如果将data_aug=data_transforms传递到数据载体,它还可以与data_augmentation一起使用。将随机种子匹配到对图像和面具的调用。
类xrv.datasets.CovariateDataset取两个数据集和两个代表标签的数组。样品将以每个站点的图像比率返回。这里的目的是模拟协变量转移,以将模型集中在不正确的功能上。然后,可以在验证数据中逆转偏移,从而导致概括性能中的灾难性故障。
比率= 0.0表示D1的图像将具有正标签比率= 0.5表示D1的图像将具有正面标签比的一半= 1.0表示D1的图像将没有正面标签
在任何比率的情况下,返回的样本数将相同。
d = xrv . datasets . CovariateDataset ( d1 = # dataset1 with a specific condition
d1_target = #target label to predict,
d2 = # dataset2 with a specific condition
d2_target = #target label to predict,
mode = "train" , # train, valid, and test
ratio = 0.9 )主TorchxrayVision论文:https://arxiv.org/abs/2111.00595
Joseph Paul Cohen, Joseph D. Viviano, Paul Bertin, Paul Morrison, Parsa Torabian, Matteo Guarrera, Matthew P Lungren, Akshay Chaudhari, Rupert Brooks, Mohammad Hashir, Hadrien Bertrand
TorchXRayVision: A library of chest X-ray datasets and models.
Medical Imaging with Deep Learning
https://github.com/mlmed/torchxrayvision, 2020
@inproceedings{Cohen2022xrv,
title = {{TorchXRayVision: A library of chest X-ray datasets and models}},
author = {Cohen, Joseph Paul and Viviano, Joseph D. and Bertin, Paul and Morrison, Paul and Torabian, Parsa and Guarrera, Matteo and Lungren, Matthew P and Chaudhari, Akshay and Brooks, Rupert and Hashir, Mohammad and Bertrand, Hadrien},
booktitle = {Medical Imaging with Deep Learning},
url = {https://github.com/mlmed/torchxrayvision},
arxivId = {2111.00595},
year = {2022}
}
以及启动图书馆开发的本文:https://arxiv.org/abs/2002.02497
Joseph Paul Cohen and Mohammad Hashir and Rupert Brooks and Hadrien Bertrand
On the limits of cross-domain generalization in automated X-ray prediction.
Medical Imaging with Deep Learning 2020 (Online: https://arxiv.org/abs/2002.02497)
@inproceedings{cohen2020limits,
title={On the limits of cross-domain generalization in automated X-ray prediction},
author={Cohen, Joseph Paul and Hashir, Mohammad and Brooks, Rupert and Bertrand, Hadrien},
booktitle={Medical Imaging with Deep Learning},
year={2020},
url={https://arxiv.org/abs/2002.02497}
}
CIFAR(加拿大高级研究所) | 蒙特利尔大学魁北克人工智能学院米拉 |
|---|---|
斯坦福大学的中心 医学和成像中的人工智能 | Carestream Health |