lingress
v3.0.7
Proyek Lingress adalah inisiatif yang bertujuan mengembangkan pipa yang ramping untuk analisis dataset resonansi magnetik nuklir (NMR), menggunakan model regresi linier univariat. Paket ini mencakup pelaksanaan analisis regresi linier melalui metode kuadrat terkecil (OLS) dan memberikan interpretasi visual dari data yang dihasilkan. Khususnya, ini termasuk nilai-p dari semua puncak NMR dalam ruang lingkup analitisnya.
Secara fungsional, program ini berusaha agar sesuai dengan model profil metabolik melalui penerapan regresi linier. Desain dan kemampuannya menyajikan alat yang kuat untuk analisis data yang mendalam dan bernuansa di ranah studi metabolisme.
pip install lingress #Example data
import numpy as np
from lingress import pickie_peak
import pandas as pd
df = pd . read_csv ( "https://raw.githubusercontent.com/aeiwz/example_data/main/dataset/Example_NMR_data.csv" )
spectra = df . iloc [:, 1 :]
ppm = spectra . columns . astype ( float ). to_list ()
#defind plot data and run UI
pickie_peak ( spectra = spectra , ppm = ppm ). run_ui ()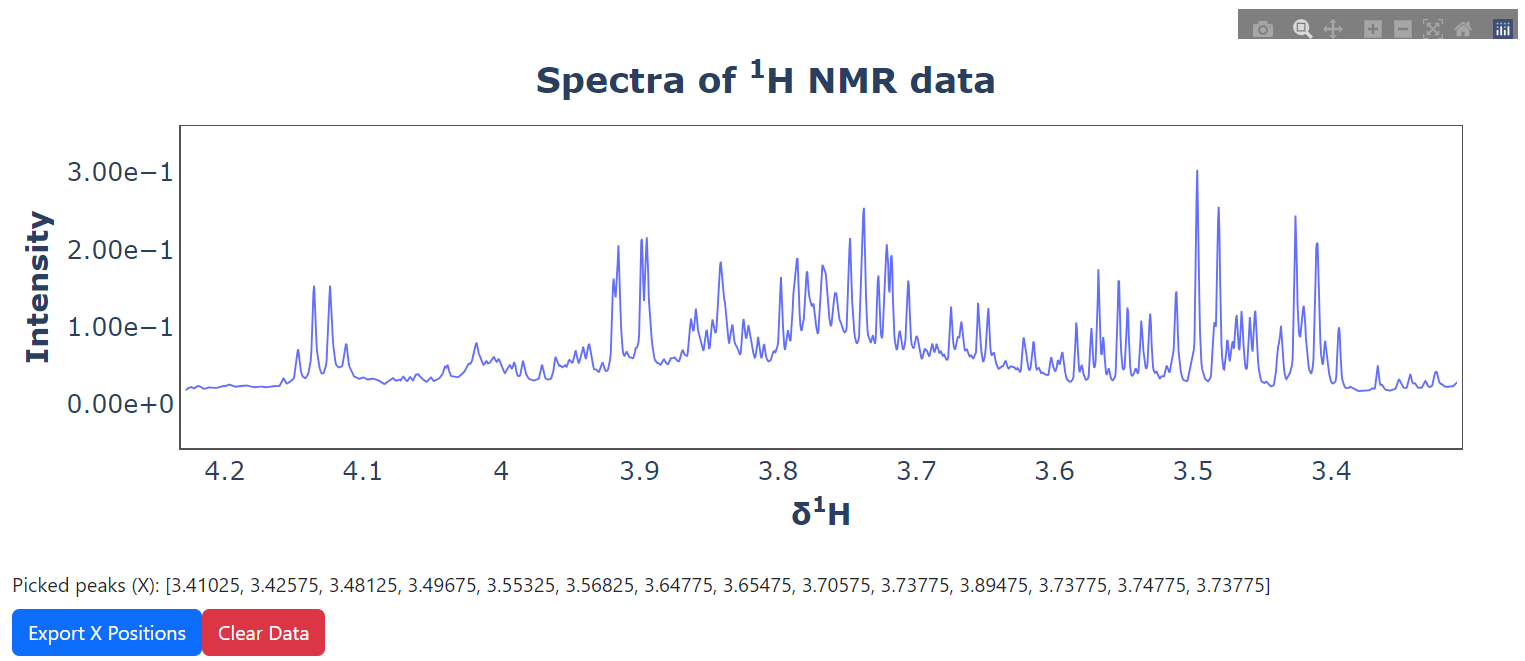
import pandas as pd
from lingress import lin_regression
df = pd . read_csv ( "https://raw.githubusercontent.com/aeiwz/example_data/main/dataset/Example_NMR_data.csv" )
X = df . iloc [:, 1 :]
ppm = spectra . columns . astype ( float ). to_list ()
y = df [ 'Group' ]
mod = lin_regression ( x = X , target = y , label = y , features_name = ppm , adj_method = 'fdr_bh' )
mod . create_dataset ()
mod . fit_model () mod . spec_uniplot ()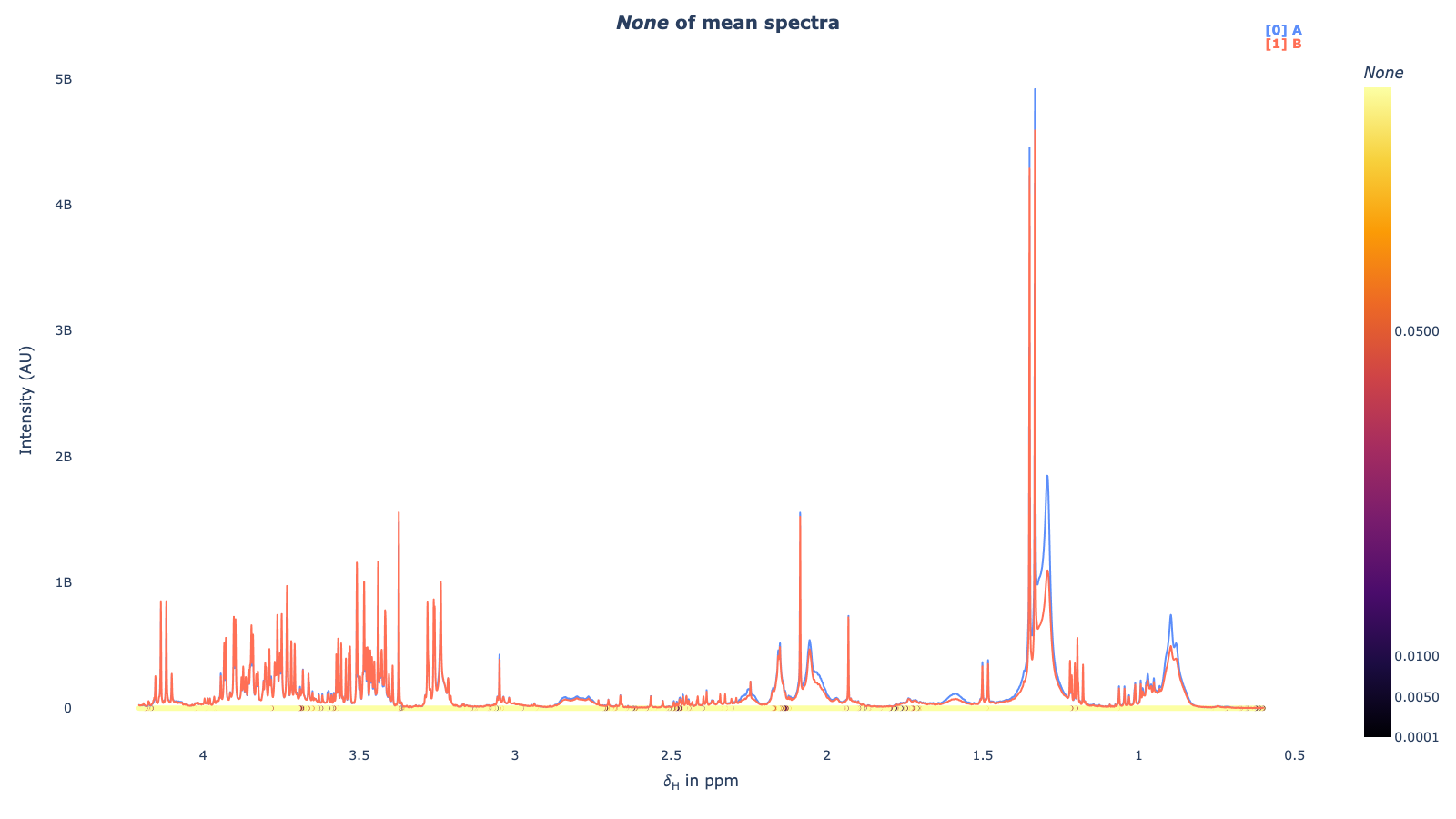
mod . volcano_plot ()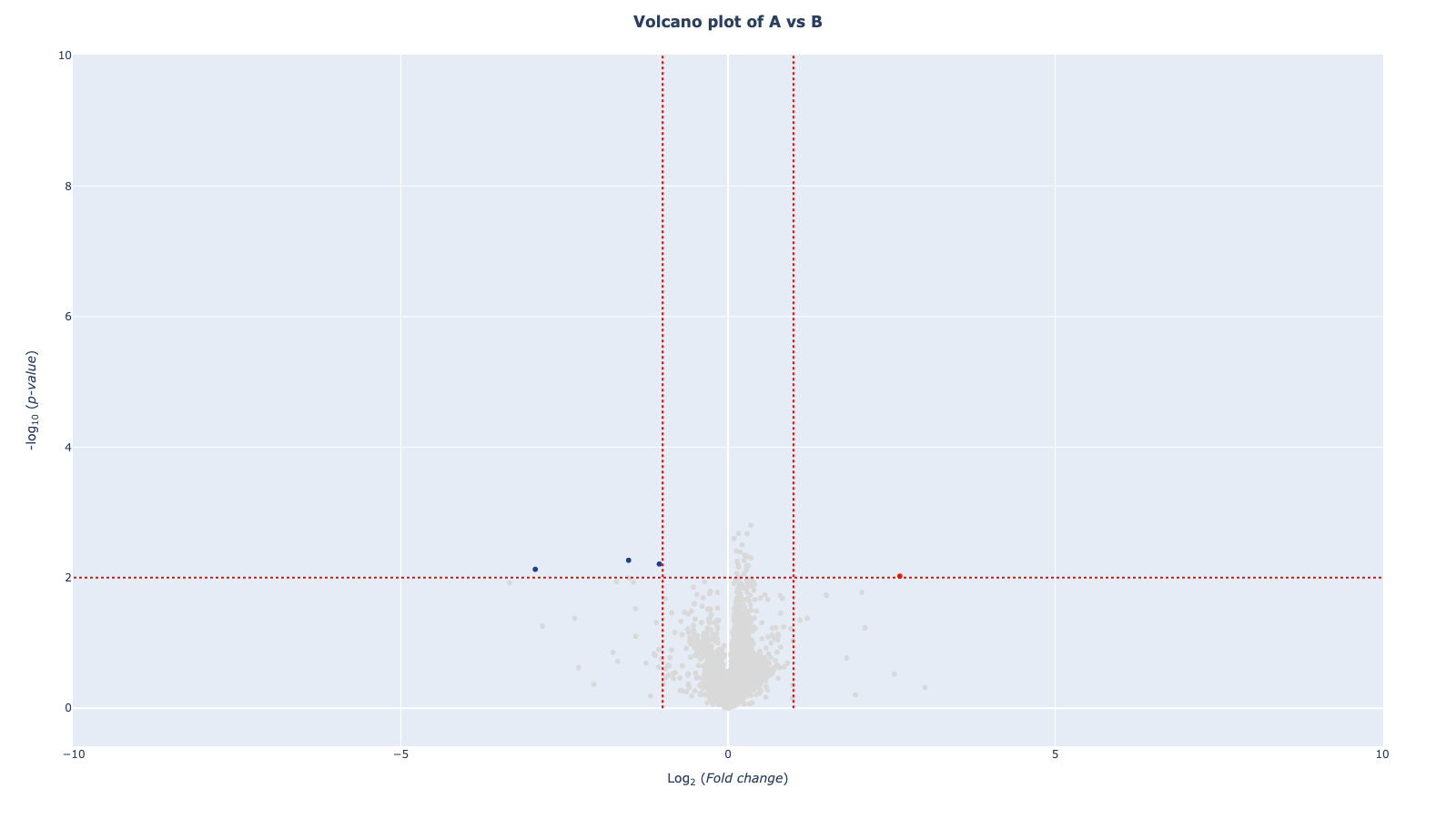
mod . resampling ( n_jobs = - 1 , n_boots = 100 , adj_method = 'fdr_bh' ) [Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers.
[Parallel(n_jobs=-1)]: Done 6 tasks | elapsed: 3.7s
[Parallel(n_jobs=-1)]: Done 60 tasks | elapsed: 6.7s
[Parallel(n_jobs=-1)]: Done 150 tasks | elapsed: 11.2s
[Parallel(n_jobs=-1)]: Done 276 tasks | elapsed: 17.8s
...
[Parallel(n_jobs=-1)]: Done 6486 tasks | elapsed: 5.6min
[Parallel(n_jobs=-1)]: Done 7188 tasks | elapsed: 6.1min
[Parallel(n_jobs=-1)]: Done 7211 out of 7211 | elapsed: 6.1min finished
mod . resampling_df ()| P-value | STD P-Value | Koefisien beta | STD Beta | Rata-rata p-value (f-test) | STD P-Value (F-Test) | Berarti r-square | STD R-Square | R2 | Penyesuaian STD R-Square | Q_Value |
|---|---|---|---|---|---|---|---|---|---|---|
| 0.60075 | 3.575454E-03 | 1.610523E-02 | 3.673194e+06 | 502596.020205 | 0.434302 | 0.276809 | 0.138650 | 0.156244 | 0.030981 | 4.012856E-03 |
| 0.60125 | 2.327687E-04 | 6.418472E-04 | 4.208365e+06 | 638734.119190 | Nan | Nan | 0.160225 | 0.175463 | 0.056503 | 3.531443E-04 |
| 0.60175 | 1.511846E-04 | 3.690541E-04 | 4.776924e+06 | 582175.023885 | 0.272894 | 0.258094 | 0.250765 | 0.204542 | 0.157111 | 2.443829E-04 |
| 0.60225 | 2.724337E-04 | 7.138873E-04 | 4.450884e+06 | 624407.676115 | 0.132108 | 0.188570 | 0.379931 | 0.198055 | 0.302422 | 4.037237E-04 |
| 0.60275 | 2.271675E-04 | 5.238926E-04 | 3.596622e+06 | 643161.588649 | 0.030732 | 0.056968 | 0.558447 | 0.158948 | 0.503253 | 3.458106E-04 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 4.20375 | 2.542707E-09 | 1.077483E-08 | 2.231841e+07 | 479783.299949 | Nan | Nan | 0.099255 | 0.130321 | -0.010838 | 4.472063E-08 |
| 4.20425 | 4.727199E-10 | 1.269310E-09 | 2.201865e+07 | 631164.491894 | 0.420162 | 0.308196 | 0.163733 | 0.184153 | 0.059199 | 1.940690e-08 |
| 4.20475 | 1.710447E-09 | 4.659603E-09 | 2.285026e+07 | 721568.566334 | Nan | Nan | 0.100927 | 0.138527 | -0.010207 | 3.595928E-08 |
| 4.20525 | 1.043658E-08 | 9.454456E-08 | 2.449345e+07 | 287615.593479 | 0.310386 | 0.301403 | 0.263740 | 0.245996 | 0.171707 | 1.084412E-07 |
| 4.20575 | 1.606948E-08 | 1.123188E-07 | 2.621135e+07 | 246414.620688 | 0.242344 | 0.257300 | 0.299881 | 0.244772 | 0.212366 | 1.457572E-07 |