El proyecto Lingress es una iniciativa destinada a desarrollar una tubería simplificada para el análisis de conjuntos de datos de resonancia magnética nuclear (RMN), utilizando un modelo de regresión lineal univariante. Este paquete abarca la ejecución del análisis de regresión lineal a través del método de mínimos cuadrados ordinarios (OLS) y proporciona interpretaciones visuales de los datos resultantes. En particular, incluye los valores p de todos los picos de RMN en su alcance analítico.
Funcionalmente, este programa se esfuerza por adaptarse a un modelo de perfiles metabólicos a través de la aplicación de la regresión lineal. Su diseño y capacidades presentan una herramienta robusta para el análisis de datos en profundidad y matizado en el ámbito de los estudios metabólicos.
pip install lingress #Example data
import numpy as np
from lingress import pickie_peak
import pandas as pd
df = pd . read_csv ( "https://raw.githubusercontent.com/aeiwz/example_data/main/dataset/Example_NMR_data.csv" )
spectra = df . iloc [:, 1 :]
ppm = spectra . columns . astype ( float ). to_list ()
#defind plot data and run UI
pickie_peak ( spectra = spectra , ppm = ppm ). run_ui ()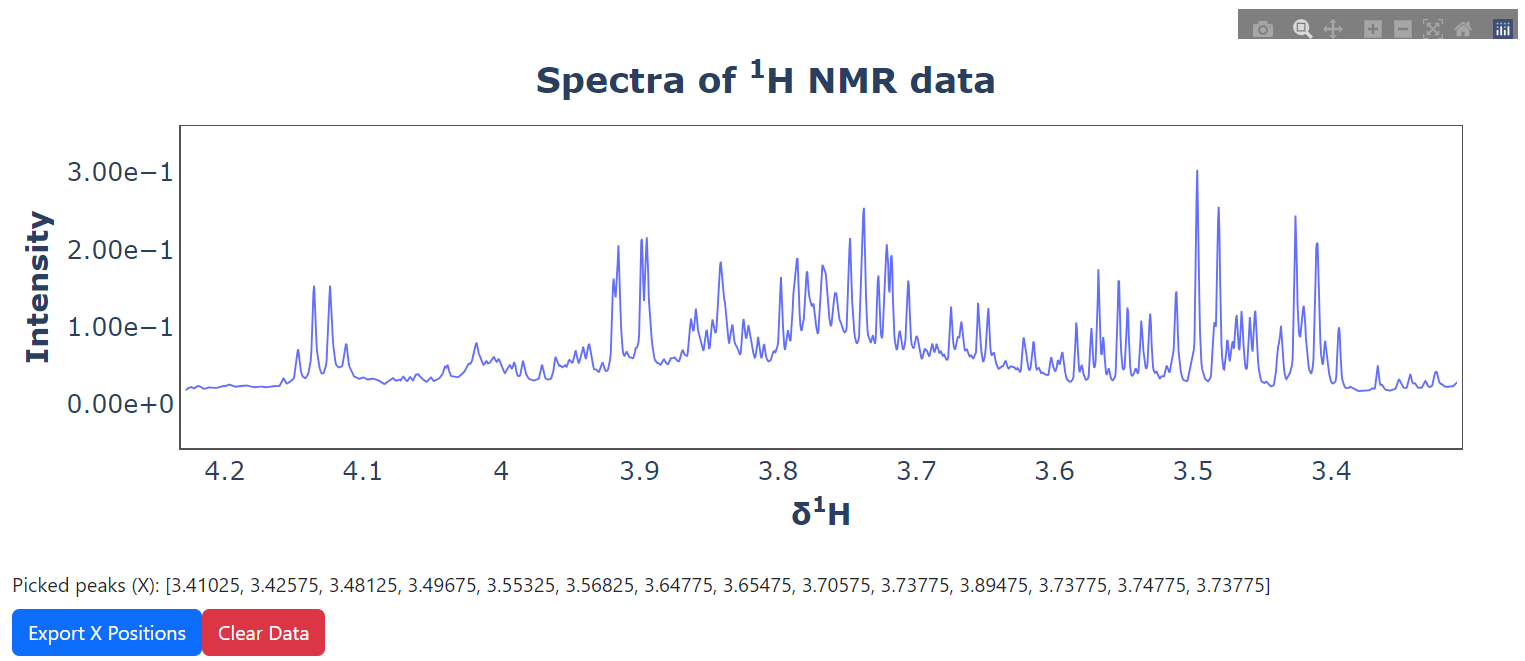
import pandas as pd
from lingress import lin_regression
df = pd . read_csv ( "https://raw.githubusercontent.com/aeiwz/example_data/main/dataset/Example_NMR_data.csv" )
X = df . iloc [:, 1 :]
ppm = spectra . columns . astype ( float ). to_list ()
y = df [ 'Group' ]
mod = lin_regression ( x = X , target = y , label = y , features_name = ppm , adj_method = 'fdr_bh' )
mod . create_dataset ()
mod . fit_model () mod . spec_uniplot ()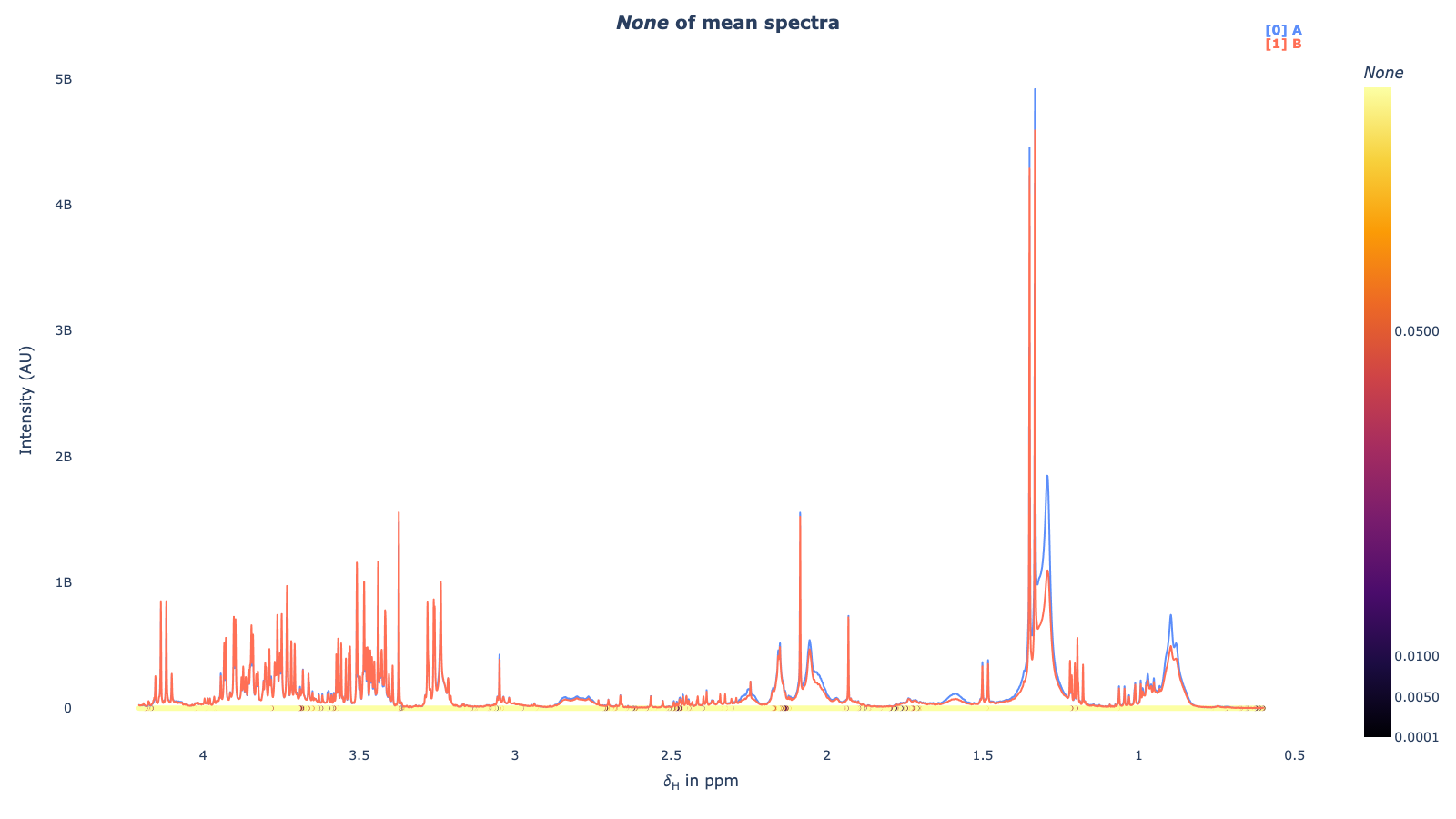
mod . volcano_plot ()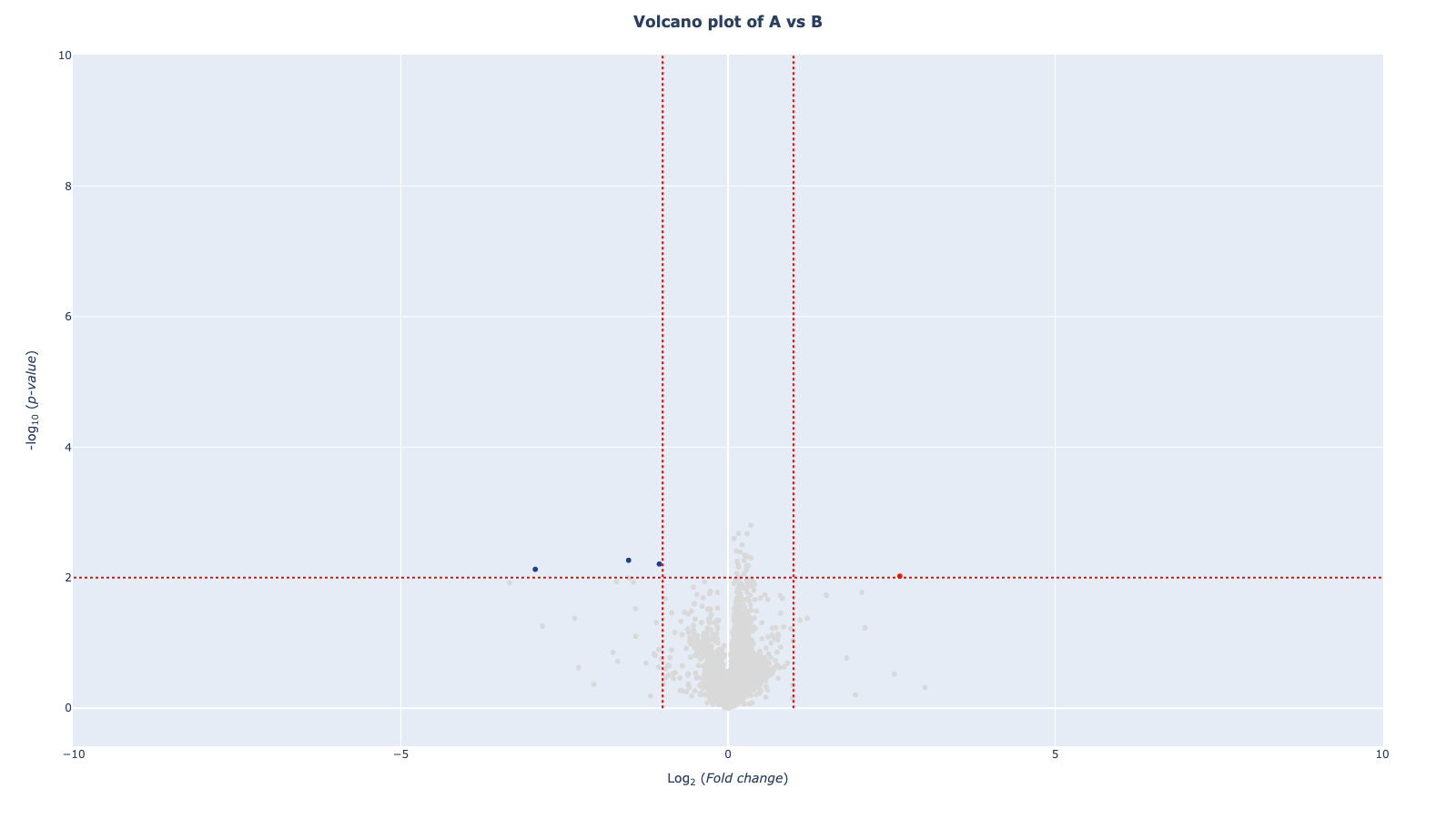
mod . resampling ( n_jobs = - 1 , n_boots = 100 , adj_method = 'fdr_bh' ) [Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers.
[Parallel(n_jobs=-1)]: Done 6 tasks | elapsed: 3.7s
[Parallel(n_jobs=-1)]: Done 60 tasks | elapsed: 6.7s
[Parallel(n_jobs=-1)]: Done 150 tasks | elapsed: 11.2s
[Parallel(n_jobs=-1)]: Done 276 tasks | elapsed: 17.8s
...
[Parallel(n_jobs=-1)]: Done 6486 tasks | elapsed: 5.6min
[Parallel(n_jobs=-1)]: Done 7188 tasks | elapsed: 6.1min
[Parallel(n_jobs=-1)]: Done 7211 out of 7211 | elapsed: 6.1min finished
mod . resampling_df ()| Valor p | Valor P de STD | Coeficiente beta | std beta | Valor P medio (prueba F) | Valor P STD (prueba F) | Medio R-cuadrado | std r-square | R2 | Ajuste de STD R-cuadrado | Q_Value |
|---|---|---|---|---|---|---|---|---|---|---|
| 0.60075 | 3.575454e-03 | 1.610523E-02 | 3.673194e+06 | 502596.020205 | 0.434302 | 0.276809 | 0.138650 | 0.156244 | 0.030981 | 4.012856E-03 |
| 0.60125 | 2.327687e-04 | 6.418472e-04 | 4.208365e+06 | 638734.119190 | Yaya | Yaya | 0.160225 | 0.175463 | 0.056503 | 3.531443e-04 |
| 0.60175 | 1.511846e-04 | 3.690541e-04 | 4.776924e+06 | 582175.023885 | 0.272894 | 0.258094 | 0.250765 | 0.204542 | 0.157111 | 2.443829e-04 |
| 0.60225 | 2.724337e-04 | 7.138873e-04 | 4.450884e+06 | 624407.676115 | 0.132108 | 0.188570 | 0.379931 | 0.198055 | 0.302422 | 4.037237e-04 |
| 0.60275 | 2.271675E-04 | 5.238926e-04 | 3.596622e+06 | 643161.588649 | 0.030732 | 0.056968 | 0.558447 | 0.158948 | 0.503253 | 3.458106E-04 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 4.20375 | 2.542707E-09 | 1.077483e-08 | 2.231841e+07 | 479783.299949 | Yaya | Yaya | 0.099255 | 0.130321 | -0.010838 | 4.472063E-08 |
| 4.20425 | 4.727199e-10 | 1.269310e-09 | 2.201865E+07 | 631164.491894 | 0.420162 | 0.308196 | 0.163733 | 0.184153 | 0.059199 | 1.940690e-08 |
| 4.20475 | 1.710447e-09 | 4.659603E-09 | 2.285026E+07 | 721568.566334 | Yaya | Yaya | 0.100927 | 0.138527 | -0.010207 | 3.595928e-08 |
| 4.20525 | 1.043658e-08 | 9.454456e-08 | 2.449345e+07 | 287615.593479 | 0.310386 | 0.301403 | 0.263740 | 0.245996 | 0.171707 | 1.084412e-07 |
| 4.20575 | 1.606948e-08 | 1.123188e-07 | 2.621135e+07 | 246414.620688 | 0.242344 | 0.257300 | 0.299881 | 0.244772 | 0.212366 | 1.457572e-07 |