skpr
v1.7.0
O SKPR é um projeto de código aberto do conjunto de experimentos para gerar e avaliar projetos ideais em R. Aqui está uma amostra do que o SKPR oferece:
# To install:
install.packages( " skpr " )
# To install the latest version from Github:
# install.packages("devtools")
devtools :: install_github( " tylermorganwall/skpr " )gen_design() gera projetos ideais de um conjunto de candidatos, dado um modelo e o número desejado de execuções.eval_design() avalia a potência parametricamente para modelos lineares, para projetos normais e de plotagem dividida.eval_design_mc() avalia poder com uma simulação de Monte Carlo, para modelos lineares lineares e generalizados. Esta função também suporta poder de cálculo para designs de plotagem dividida usando o REML.eval_design_survival_mc() Avalia energia com uma simulação de Monte Carlo, permitindo que o usuário especifique um ponto em que os dados são censurados.eval_design_custom_mc() permite que o usuário importe suas próprias bibliotecas e use a estrutura Monte Carlo fornecida pelo SKPR para calcular a energia.calculate_power_curves() fornece uma interface para automatizar a geração e avaliação de projetos para criar energia versus tamanho de amostra e curvas de tamanho de efeito.skprGUI() abre a GUI em RStudio ou um navegador externo.Se a adição, o pacote oferece duas funções para gerar parcelas comuns relacionadas a projetos:
plot_correlations() gera um mapa de cores de correlações entre variáveis.plot_fds() gera a fração do gráfico de espaço de design para um determinado design.## skprgui
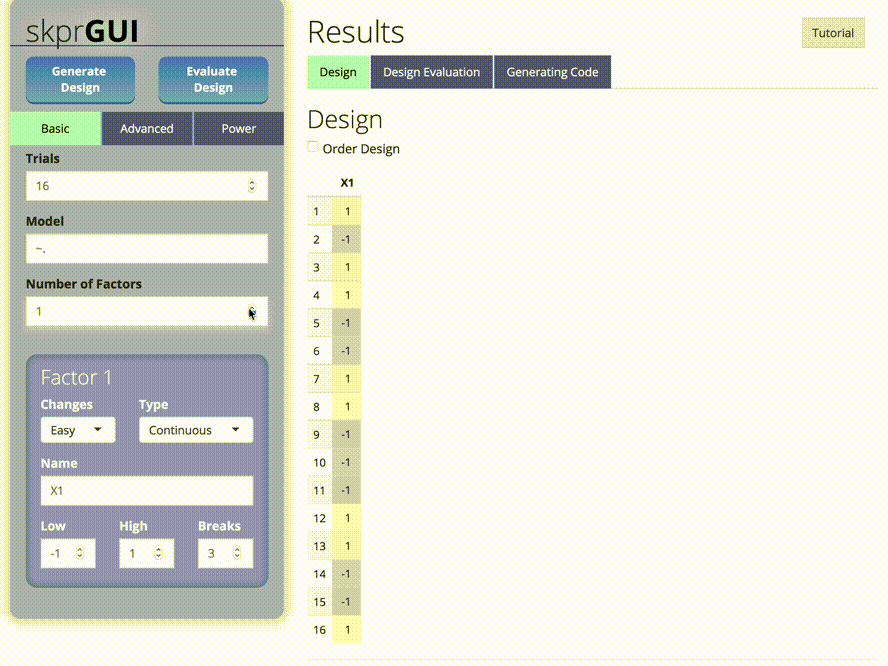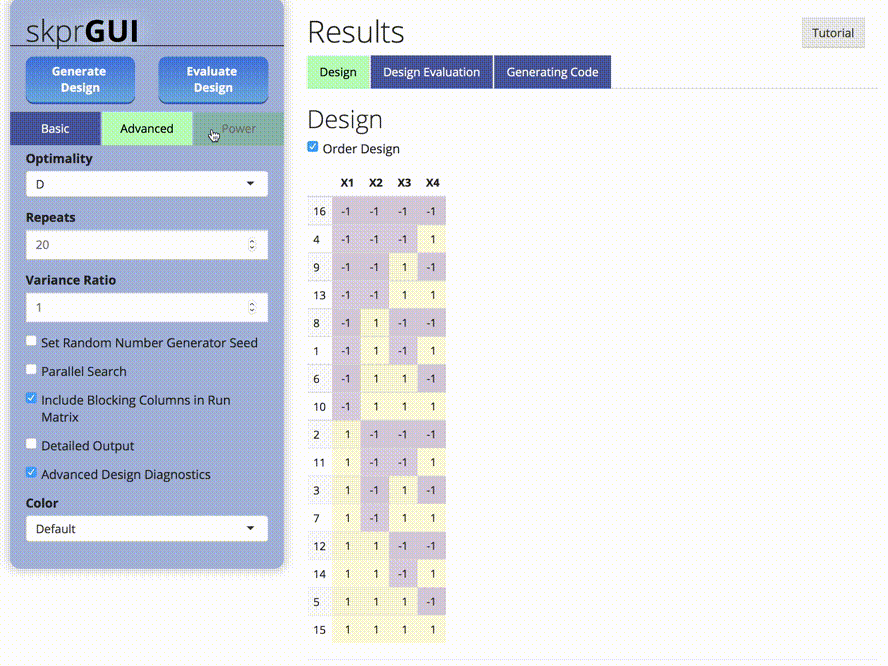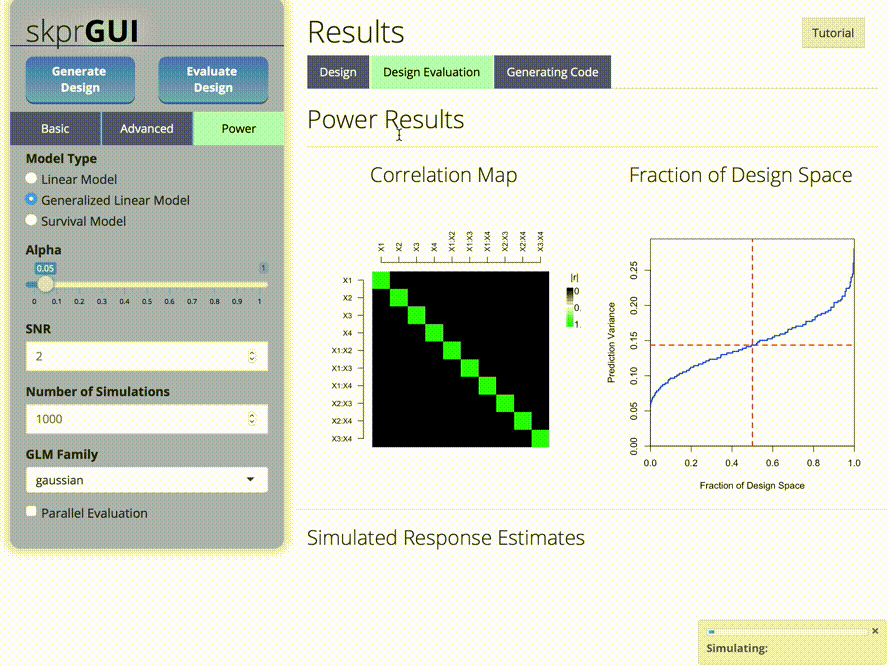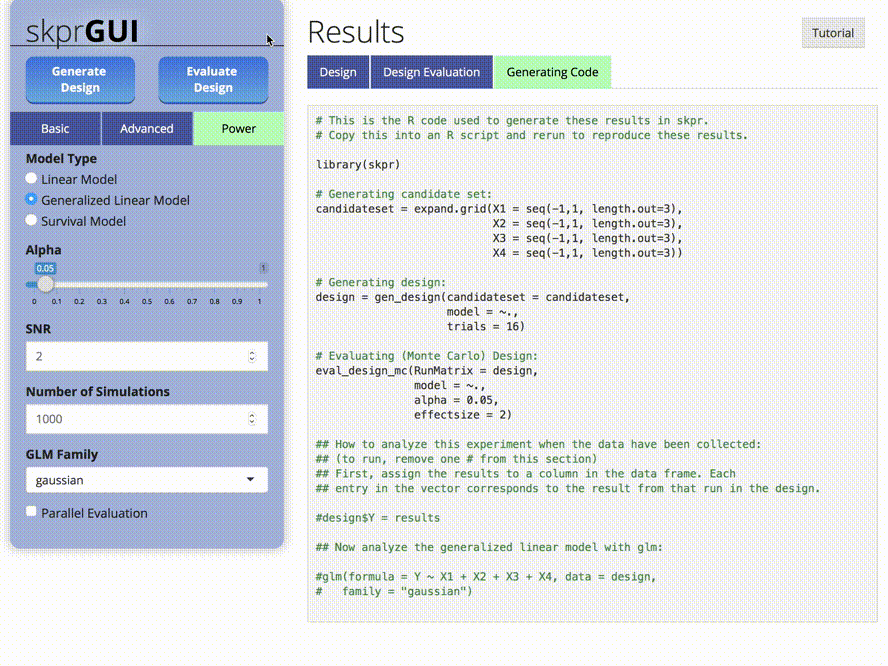
skprGUI() fornece uma interface gráfica do usuário para acessar todos os principais recursos do SKPR. Um tutorial interativo é fornecido para familiarizar o usuário com a funcionalidade disponível. Tipo skprGUI() para começar. Capturas de tela:
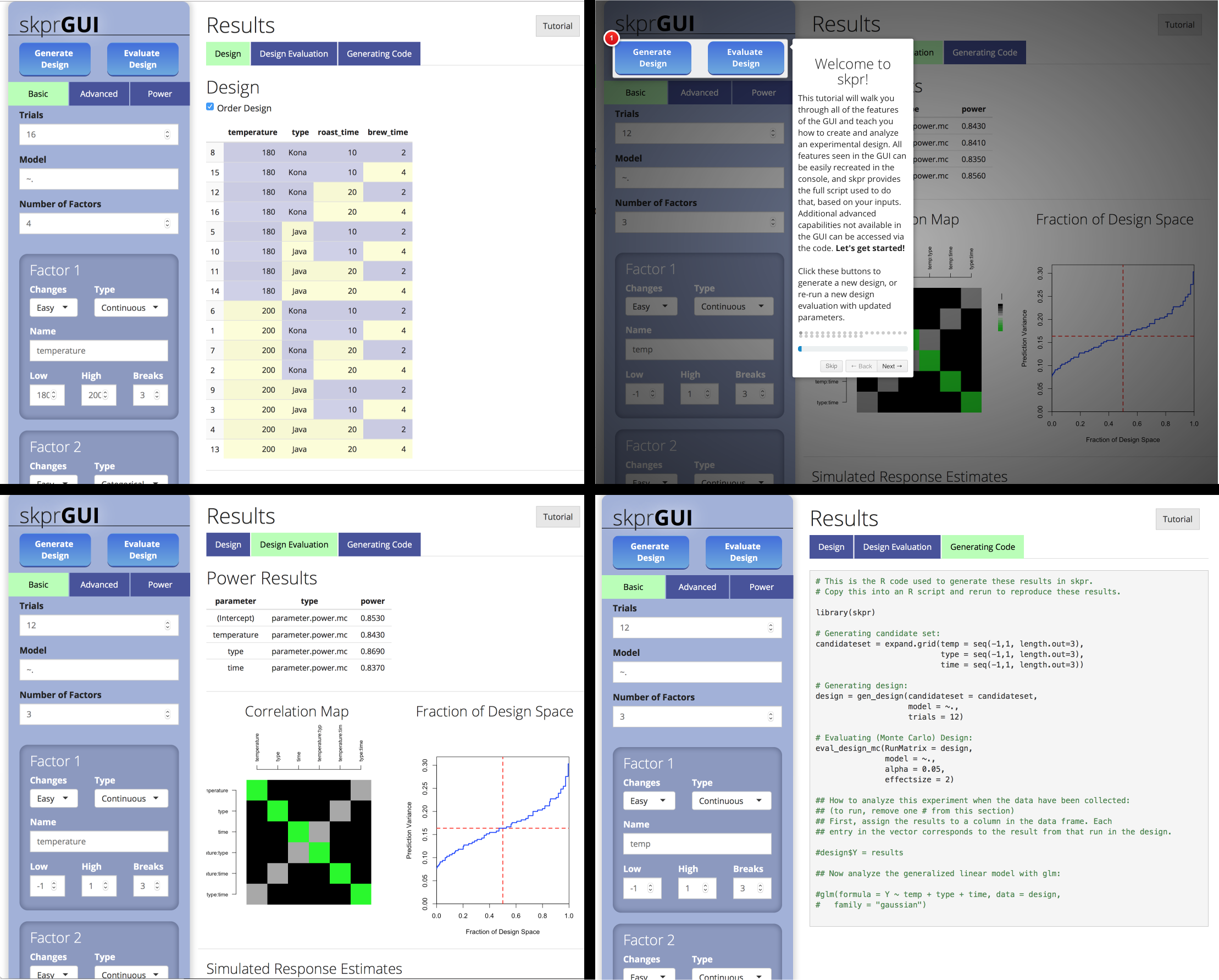
library( skpr )
# Generate a candidate set of all potential design points to be considered in the experiment
# The hypothetical experiment is determining what affects the caffeine content in coffee
candidate_set = expand.grid( temp = c( 80 , 90 , 100 ),
type = c( " Kona " , " Java " ),
beansize = c( " Large " , " Medium " , " Small " ))
candidate_set
# > temp type beansize
# > 1 80 Kona Large
# > 2 90 Kona Large
# > 3 100 Kona Large
# > 4 80 Java Large
# > 5 90 Java Large
# > 6 100 Java Large
# > 7 80 Kona Medium
# > 8 90 Kona Medium
# > 9 100 Kona Medium
# > 10 80 Java Medium
# > 11 90 Java Medium
# > 12 100 Java Medium
# > 13 80 Kona Small
# > 14 90 Kona Small
# > 15 100 Kona Small
# > 16 80 Java Small
# > 17 90 Java Small
# > 18 100 Java Small
# Generate the design (default D-optimal)
design = gen_design( candidateset = candidate_set ,
model = ~ temp + type + beansize ,
trials = 12 )
design
# > temp type beansize
# > 1 80 Java Medium
# > 2 100 Java Large
# > 3 100 Java Small
# > 4 80 Java Large
# > 5 80 Kona Medium
# > 6 80 Kona Small
# > 7 100 Kona Small
# > 8 100 Kona Medium
# > 9 80 Kona Large
# > 10 100 Java Medium
# > 11 100 Kona Large
# > 12 80 Java Small
# Evaluate power for the design with an allowable type-I error of 5% (default)
eval_design( design )
# > parameter type power
# > 1 (Intercept) effect.power 0.8424665
# > 2 temp effect.power 0.8424665
# > 3 type effect.power 0.8424665
# > 4 beansize effect.power 0.5165386
# > 5 (Intercept) parameter.power 0.8424665
# > 6 temp parameter.power 0.8424665
# > 7 type1 parameter.power 0.8424665
# > 8 beansize1 parameter.power 0.5593966
# > 9 beansize2 parameter.power 0.5593966
# > ============Evaluation Info============
# > * Alpha = 0.05 * Trials = 12 * Blocked = FALSE
# > * Evaluating Model = ~temp + type + beansize
# > * Anticipated Coefficients = c(1, 1, 1, 1, -1)
# > * Contrasts = `contr.sum`
# > * Parameter Analysis Method = `lm(...)`
# > * Effect Analysis Method = `car::Anova(fit, type = "III")`
# Evaluate power for the design using a Monte Carlo simulation.
# Here, we set the effect size (here, the signal-to-noise ratio) to 1.5.
eval_design_mc( design , effectsize = 1.5 )
# > parameter type power
# > 1 (Intercept) effect.power.mc 0.600
# > 2 temp effect.power.mc 0.612
# > 3 type effect.power.mc 0.610
# > 4 beansize effect.power.mc 0.316
# > 5 (Intercept) parameter.power.mc 0.600
# > 6 temp parameter.power.mc 0.612
# > 7 type1 parameter.power.mc 0.610
# > 8 beansize1 parameter.power.mc 0.359
# > 9 beansize2 parameter.power.mc 0.354
# > ===========Evaluation Info============
# > * Alpha = 0.05 * Trials = 12 * Blocked = FALSE
# > * Evaluating Model = ~temp + type + beansize
# > * Anticipated Coefficients = c(0.750, 0.750, 0.750, 0.750, -0.750)
# > * Contrasts = `contr.sum`
# > * Parameter Analysis Method = `lm(...)`
# > * Effect Analysis Method = `car::Anova(fit, type = "III")`
# Evaluate power for the design using a Monte Carlo simulation, for a non-normal response.
# Here, we also increase the number of simululations to improve the precision of the results.
eval_design_mc( design , nsim = 5000 , glmfamily = " poisson " , effectsize = c( 2 , 6 ))
# > parameter type power
# > 1 (Intercept) effect.power.mc 0.9968
# > 2 temp effect.power.mc 0.9826
# > 3 type effect.power.mc 0.9832
# > 4 beansize effect.power.mc 0.8502
# > 5 (Intercept) parameter.power.mc 0.9968
# > 6 temp parameter.power.mc 0.9826
# > 7 type1 parameter.power.mc 0.9832
# > 8 beansize1 parameter.power.mc 0.8842
# > 9 beansize2 parameter.power.mc 0.7052
# > ============Evaluation Info============
# > * Alpha = 0.05 * Trials = 12 * Blocked = FALSE
# > * Evaluating Model = ~temp + type + beansize
# > * Anticipated Coefficients = c(1.242, 0.549, 0.549, 0.549, -0.549)
# > * Contrasts = `contr.sum`
# > * Parameter Analysis Method = `glm(..., family = "poisson")`
# > * Effect Analysis Method = `car::Anova(fit, type = "III")`
# skpr was designed to operate with the pipe (|>) in mind.
# Here is an example of an entire design of experiments analysis in three lines:
expand.grid( temp = c( 80 , 90 , 100 ), type = c( " Kona " , " Java " ), beansize = c( " Large " , " Medium " , " Small " )) | >
gen_design( model = ~ temp + type + beansize + beansize : type + I( temp ^ 2 ), trials = 24 , optimality = " I " ) | >
eval_design_mc( detailedoutput = TRUE )
# > parameter type power anticoef alpha glmfamily trials
# > 1 (Intercept) effect.power.mc 0.912 NA 0.05 gaussian 24
# > 2 temp effect.power.mc 0.927 NA 0.05 gaussian 24
# > 3 type effect.power.mc 0.997 NA 0.05 gaussian 24
# > 4 beansize effect.power.mc 0.935 NA 0.05 gaussian 24
# > 5 I(temp^2) effect.power.mc 0.637 NA 0.05 gaussian 24
# > 6 type:beansize effect.power.mc 0.913 NA 0.05 gaussian 24
# > 7 (Intercept) parameter.power.mc 0.912 1 0.05 gaussian 24
# > 8 temp parameter.power.mc 0.927 1 0.05 gaussian 24
# > 9 type1 parameter.power.mc 0.997 1 0.05 gaussian 24
# > 10 beansize1 parameter.power.mc 0.917 1 0.05 gaussian 24
# > 11 beansize2 parameter.power.mc 0.913 -1 0.05 gaussian 24
# > 12 I(temp^2) parameter.power.mc 0.637 1 0.05 gaussian 24
# > 13 type1:beansize1 parameter.power.mc 0.899 1 0.05 gaussian 24
# > 14 type1:beansize2 parameter.power.mc 0.902 -1 0.05 gaussian 24
# > nsim blocking error_adjusted_alpha power_lcb power_ucb
# > 1 1000 FALSE 0.05 0.8927052 0.9288249
# > 2 1000 FALSE 0.05 0.9090858 0.9423464
# > 3 1000 FALSE 0.05 0.9912580 0.9993809
# > 4 1000 FALSE 0.05 0.9178989 0.9494797
# > 5 1000 FALSE 0.05 0.6063275 0.6668632
# > 6 1000 FALSE 0.05 0.8937921 0.9297315
# > 7 1000 FALSE 0.05 0.8927052 0.9288249
# > 8 1000 FALSE 0.05 0.9090858 0.9423464
# > 9 1000 FALSE 0.05 0.9912580 0.9993809
# > 10 1000 FALSE 0.05 0.8981467 0.9333511
# > 11 1000 FALSE 0.05 0.8937921 0.9297315
# > 12 1000 FALSE 0.05 0.6063275 0.6668632
# > 13 1000 FALSE 0.05 0.8786332 0.9169799
# > 14 1000 FALSE 0.05 0.8818715 0.9197225
# > =========================================================Evaluation Info==========================================================
# > * Alpha = 0.05 * Trials = 24 * Blocked = FALSE
# > * Evaluating Model = ~temp + type + beansize + type:beansize + I(temp^2)
# > * Anticipated Coefficients = c(1, 1, 1, 1, -1, 1, 1, -1)
# > * Contrasts = `contr.sum`
# > * Parameter Analysis Method = `lm(...)`
# > * Effect Analysis Method = `car::Anova(fit, type = "III")`
# > * MC Power CI Confidence = 95%