skpr
v1.7.0
SKPR adalah desain open source dari eksperimen suite untuk menghasilkan dan mengevaluasi desain optimal di R. Berikut ini adalah contoh dari apa yang ditawarkan SKPR:
# To install:
install.packages( " skpr " )
# To install the latest version from Github:
# install.packages("devtools")
devtools :: install_github( " tylermorganwall/skpr " )gen_design() menghasilkan desain optimal dari kandidat set, diberi model dan jumlah berjalan yang diinginkan.eval_design() mengevaluasi daya secara parametrik untuk model linier, untuk desain normal dan split-plot.eval_design_mc() mengevaluasi daya dengan simulasi Monte Carlo, untuk model linier linier dan umum. Fungsi ini juga mendukung daya penghitungan untuk desain split-plot menggunakan REML.eval_design_survival_mc() mengevaluasi daya dengan simulasi Monte Carlo, yang memungkinkan pengguna untuk menentukan titik di mana data disensor.eval_design_custom_mc() memungkinkan pengguna untuk mengimpor perpustakaan mereka sendiri dan menggunakan kerangka kerja Monte Carlo yang disediakan oleh SKPR untuk menghitung daya.calculate_power_curves() menyediakan antarmuka untuk mengotomatisasi generasi dan evaluasi desain untuk membuat daya versus ukuran sampel dan kurva ukuran efek.skprGUI() membuka GUI baik di rstudio atau browser eksternal.Jika penambahan, paket ini menawarkan dua fungsi untuk menghasilkan plot umum yang terkait dengan desain:
plot_correlations() menghasilkan peta warna korelasi antar variabel.plot_fds() menghasilkan fraksi plot ruang desain untuk desain yang diberikan.## skprgui
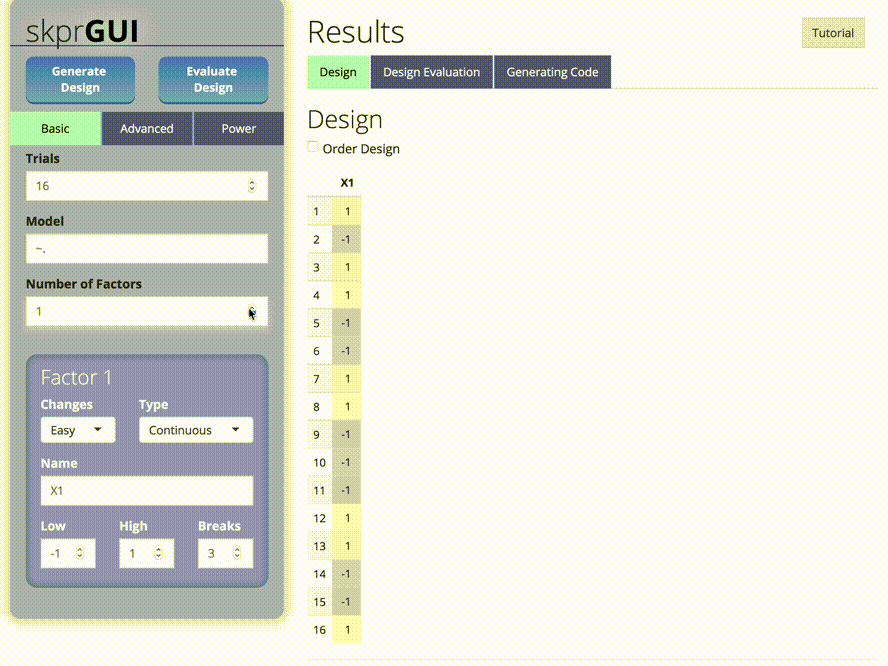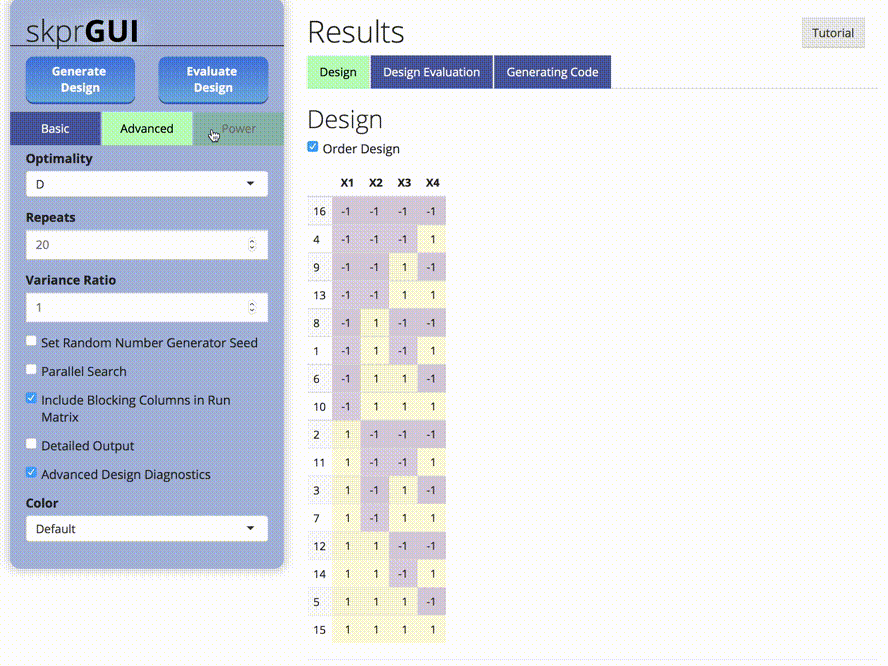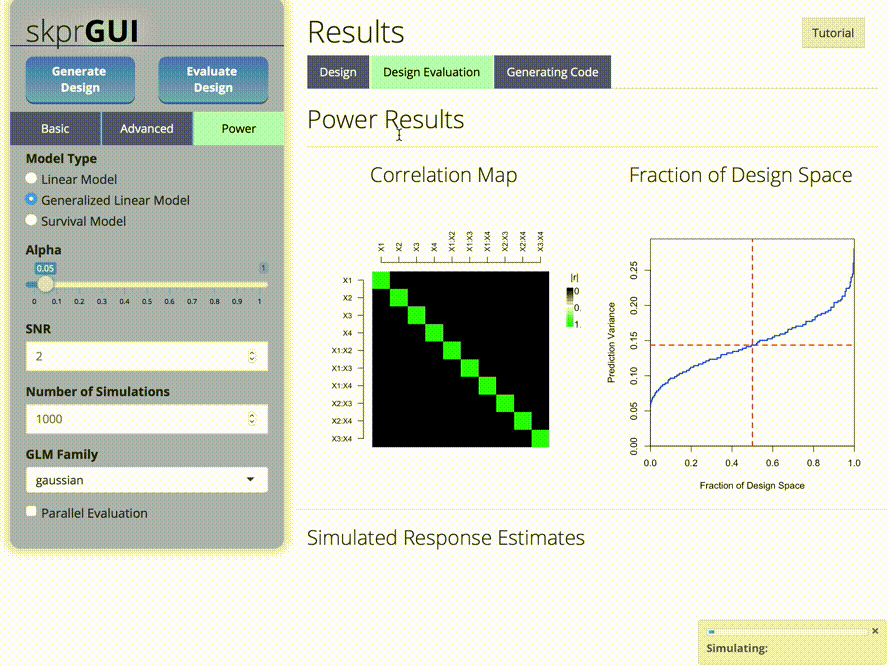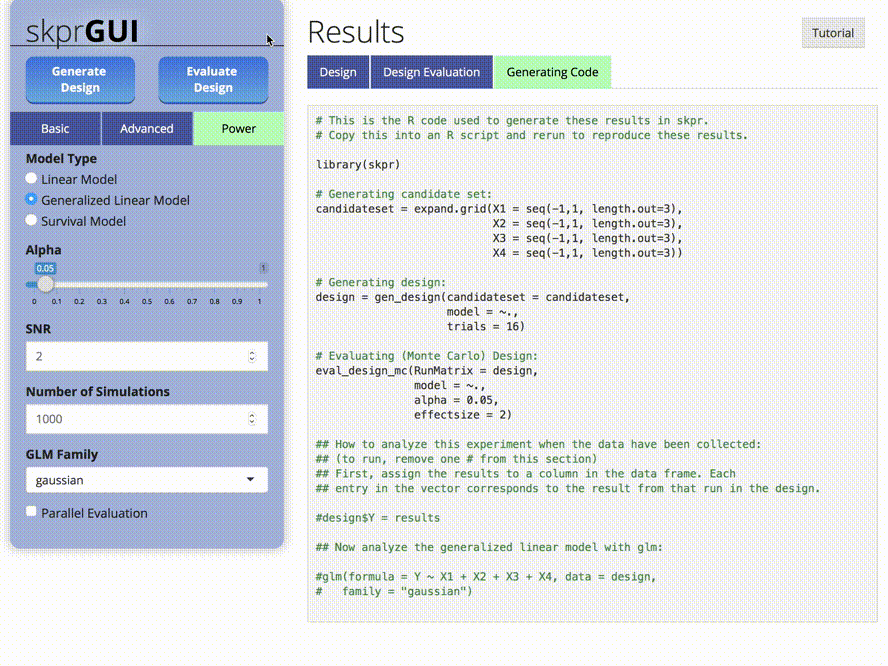
skprGUI() menyediakan antarmuka pengguna grafis untuk mengakses semua fitur utama SKPR. Tutorial interaktif disediakan untuk membiasakan pengguna dengan fungsionalitas yang tersedia. Ketik skprGUI() untuk memulai. Tangkapan layar:
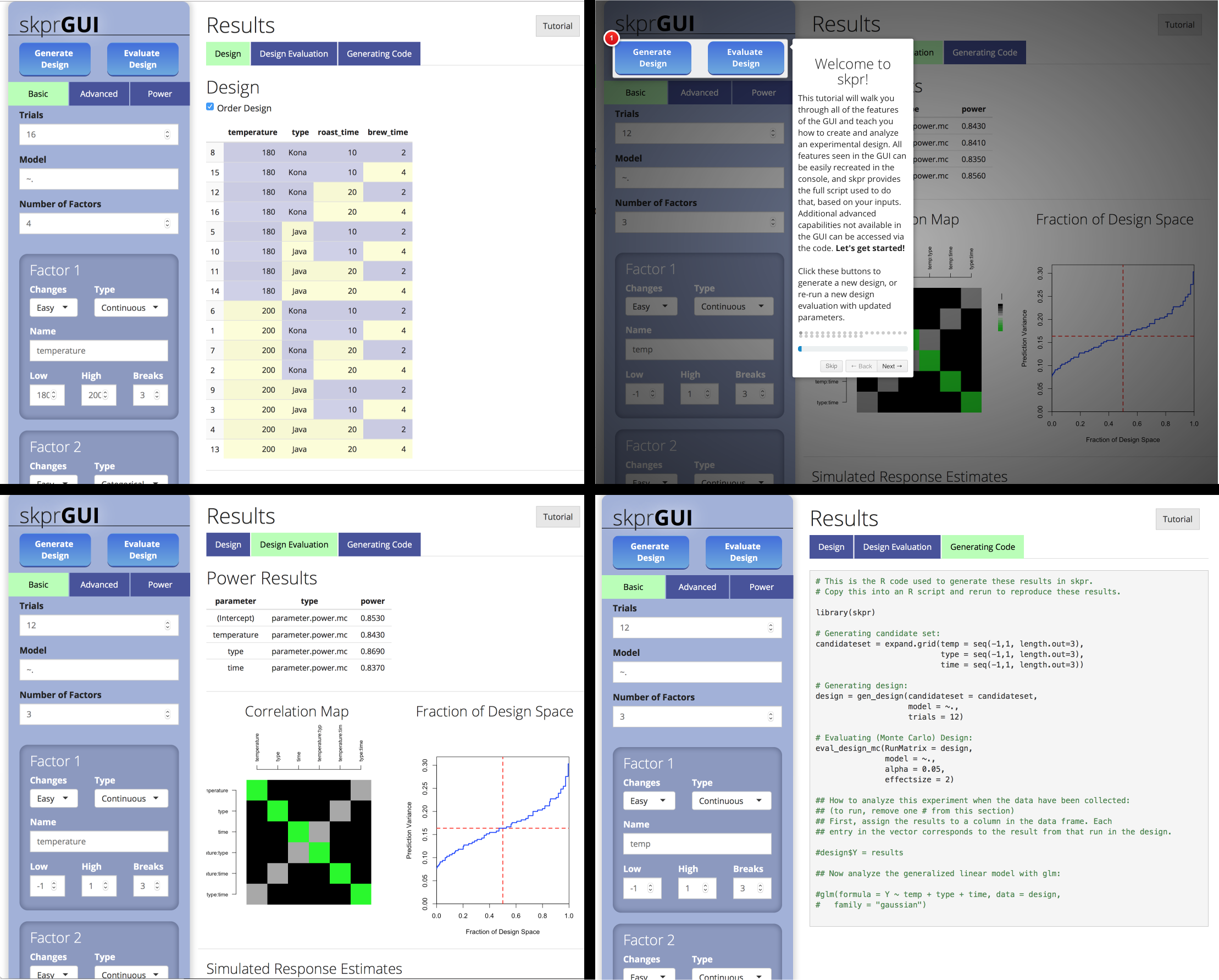
library( skpr )
# Generate a candidate set of all potential design points to be considered in the experiment
# The hypothetical experiment is determining what affects the caffeine content in coffee
candidate_set = expand.grid( temp = c( 80 , 90 , 100 ),
type = c( " Kona " , " Java " ),
beansize = c( " Large " , " Medium " , " Small " ))
candidate_set
# > temp type beansize
# > 1 80 Kona Large
# > 2 90 Kona Large
# > 3 100 Kona Large
# > 4 80 Java Large
# > 5 90 Java Large
# > 6 100 Java Large
# > 7 80 Kona Medium
# > 8 90 Kona Medium
# > 9 100 Kona Medium
# > 10 80 Java Medium
# > 11 90 Java Medium
# > 12 100 Java Medium
# > 13 80 Kona Small
# > 14 90 Kona Small
# > 15 100 Kona Small
# > 16 80 Java Small
# > 17 90 Java Small
# > 18 100 Java Small
# Generate the design (default D-optimal)
design = gen_design( candidateset = candidate_set ,
model = ~ temp + type + beansize ,
trials = 12 )
design
# > temp type beansize
# > 1 80 Java Medium
# > 2 100 Java Large
# > 3 100 Java Small
# > 4 80 Java Large
# > 5 80 Kona Medium
# > 6 80 Kona Small
# > 7 100 Kona Small
# > 8 100 Kona Medium
# > 9 80 Kona Large
# > 10 100 Java Medium
# > 11 100 Kona Large
# > 12 80 Java Small
# Evaluate power for the design with an allowable type-I error of 5% (default)
eval_design( design )
# > parameter type power
# > 1 (Intercept) effect.power 0.8424665
# > 2 temp effect.power 0.8424665
# > 3 type effect.power 0.8424665
# > 4 beansize effect.power 0.5165386
# > 5 (Intercept) parameter.power 0.8424665
# > 6 temp parameter.power 0.8424665
# > 7 type1 parameter.power 0.8424665
# > 8 beansize1 parameter.power 0.5593966
# > 9 beansize2 parameter.power 0.5593966
# > ============Evaluation Info============
# > * Alpha = 0.05 * Trials = 12 * Blocked = FALSE
# > * Evaluating Model = ~temp + type + beansize
# > * Anticipated Coefficients = c(1, 1, 1, 1, -1)
# > * Contrasts = `contr.sum`
# > * Parameter Analysis Method = `lm(...)`
# > * Effect Analysis Method = `car::Anova(fit, type = "III")`
# Evaluate power for the design using a Monte Carlo simulation.
# Here, we set the effect size (here, the signal-to-noise ratio) to 1.5.
eval_design_mc( design , effectsize = 1.5 )
# > parameter type power
# > 1 (Intercept) effect.power.mc 0.600
# > 2 temp effect.power.mc 0.612
# > 3 type effect.power.mc 0.610
# > 4 beansize effect.power.mc 0.316
# > 5 (Intercept) parameter.power.mc 0.600
# > 6 temp parameter.power.mc 0.612
# > 7 type1 parameter.power.mc 0.610
# > 8 beansize1 parameter.power.mc 0.359
# > 9 beansize2 parameter.power.mc 0.354
# > ===========Evaluation Info============
# > * Alpha = 0.05 * Trials = 12 * Blocked = FALSE
# > * Evaluating Model = ~temp + type + beansize
# > * Anticipated Coefficients = c(0.750, 0.750, 0.750, 0.750, -0.750)
# > * Contrasts = `contr.sum`
# > * Parameter Analysis Method = `lm(...)`
# > * Effect Analysis Method = `car::Anova(fit, type = "III")`
# Evaluate power for the design using a Monte Carlo simulation, for a non-normal response.
# Here, we also increase the number of simululations to improve the precision of the results.
eval_design_mc( design , nsim = 5000 , glmfamily = " poisson " , effectsize = c( 2 , 6 ))
# > parameter type power
# > 1 (Intercept) effect.power.mc 0.9968
# > 2 temp effect.power.mc 0.9826
# > 3 type effect.power.mc 0.9832
# > 4 beansize effect.power.mc 0.8502
# > 5 (Intercept) parameter.power.mc 0.9968
# > 6 temp parameter.power.mc 0.9826
# > 7 type1 parameter.power.mc 0.9832
# > 8 beansize1 parameter.power.mc 0.8842
# > 9 beansize2 parameter.power.mc 0.7052
# > ============Evaluation Info============
# > * Alpha = 0.05 * Trials = 12 * Blocked = FALSE
# > * Evaluating Model = ~temp + type + beansize
# > * Anticipated Coefficients = c(1.242, 0.549, 0.549, 0.549, -0.549)
# > * Contrasts = `contr.sum`
# > * Parameter Analysis Method = `glm(..., family = "poisson")`
# > * Effect Analysis Method = `car::Anova(fit, type = "III")`
# skpr was designed to operate with the pipe (|>) in mind.
# Here is an example of an entire design of experiments analysis in three lines:
expand.grid( temp = c( 80 , 90 , 100 ), type = c( " Kona " , " Java " ), beansize = c( " Large " , " Medium " , " Small " )) | >
gen_design( model = ~ temp + type + beansize + beansize : type + I( temp ^ 2 ), trials = 24 , optimality = " I " ) | >
eval_design_mc( detailedoutput = TRUE )
# > parameter type power anticoef alpha glmfamily trials
# > 1 (Intercept) effect.power.mc 0.912 NA 0.05 gaussian 24
# > 2 temp effect.power.mc 0.927 NA 0.05 gaussian 24
# > 3 type effect.power.mc 0.997 NA 0.05 gaussian 24
# > 4 beansize effect.power.mc 0.935 NA 0.05 gaussian 24
# > 5 I(temp^2) effect.power.mc 0.637 NA 0.05 gaussian 24
# > 6 type:beansize effect.power.mc 0.913 NA 0.05 gaussian 24
# > 7 (Intercept) parameter.power.mc 0.912 1 0.05 gaussian 24
# > 8 temp parameter.power.mc 0.927 1 0.05 gaussian 24
# > 9 type1 parameter.power.mc 0.997 1 0.05 gaussian 24
# > 10 beansize1 parameter.power.mc 0.917 1 0.05 gaussian 24
# > 11 beansize2 parameter.power.mc 0.913 -1 0.05 gaussian 24
# > 12 I(temp^2) parameter.power.mc 0.637 1 0.05 gaussian 24
# > 13 type1:beansize1 parameter.power.mc 0.899 1 0.05 gaussian 24
# > 14 type1:beansize2 parameter.power.mc 0.902 -1 0.05 gaussian 24
# > nsim blocking error_adjusted_alpha power_lcb power_ucb
# > 1 1000 FALSE 0.05 0.8927052 0.9288249
# > 2 1000 FALSE 0.05 0.9090858 0.9423464
# > 3 1000 FALSE 0.05 0.9912580 0.9993809
# > 4 1000 FALSE 0.05 0.9178989 0.9494797
# > 5 1000 FALSE 0.05 0.6063275 0.6668632
# > 6 1000 FALSE 0.05 0.8937921 0.9297315
# > 7 1000 FALSE 0.05 0.8927052 0.9288249
# > 8 1000 FALSE 0.05 0.9090858 0.9423464
# > 9 1000 FALSE 0.05 0.9912580 0.9993809
# > 10 1000 FALSE 0.05 0.8981467 0.9333511
# > 11 1000 FALSE 0.05 0.8937921 0.9297315
# > 12 1000 FALSE 0.05 0.6063275 0.6668632
# > 13 1000 FALSE 0.05 0.8786332 0.9169799
# > 14 1000 FALSE 0.05 0.8818715 0.9197225
# > =========================================================Evaluation Info==========================================================
# > * Alpha = 0.05 * Trials = 24 * Blocked = FALSE
# > * Evaluating Model = ~temp + type + beansize + type:beansize + I(temp^2)
# > * Anticipated Coefficients = c(1, 1, 1, 1, -1, 1, 1, -1)
# > * Contrasts = `contr.sum`
# > * Parameter Analysis Method = `lm(...)`
# > * Effect Analysis Method = `car::Anova(fit, type = "III")`
# > * MC Power CI Confidence = 95%