Non-targeted metabolomics plays a crucial role in the wave of precision medicine and biomarker discovery. However, the identification of compounds remains challenging due to the incompleteness of the existing spectrum reference library. To solve this problem, the German Federal Institute of Materials Research and Testing (BAM) and a research team from the Free University of Berlin jointly developed FIORA, an open source graph neural network (GNN), designed to simulate the process of tandem mass spectrometry to help improve the accuracy of mass spectrometry recognition.
The core of the FIORA model is that it uses local neighborhood information of bonds in molecules to learn the breaking patterns of compounds, thereby deriving the probability of fragmented ions. Compared with the traditional fragmentation algorithms ICEBERG and CFM-ID, FIORA performs excellent in quality prediction and can predict other features such as retention time (RT) and collision cross-section (CCS). This pioneering research result was published in Nature Communications on March 7, 2025.
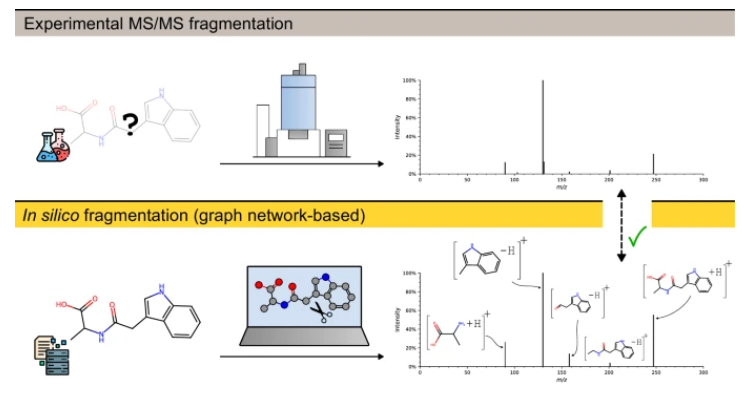
FIORA’s design makes full use of high-performance GPUs, quickly validate putative compound annotations, and significantly expands the spectral reference library with high-quality predictions. This progress is of great significance to promote research on non-targeted metabolomics, especially when analyzing unknown compounds. Research in this field has been slow in the past decade due to the scarcity of high-quality reference spectrum. For example, the 2016 CASMI Challenge showed that the recall rate of computer simulation methods was only 34%, while it was not even 30% in 2022. This shows the urgent need for a new solution.
FIORA is unique in that it can independently evaluate bond dissociation events based on the local structure of each compound. This method simulates the physical fragmentation process in mass spectrometry more directly than many existing algorithms. Furthermore, FIORA not only performs well when targeting similar compounds, but also its ability to promote unfamiliar structures is impressive.
To ensure its effectiveness, FIORA tested on multiple data sets and the results showed that the median similarity between its predicted mass spectra and reference spectrum reached more than 0.8, and in some cases, 10% to 49% higher than the competition algorithm. In addition, FIORA's modular design allows it to adapt flexibly to different predictive goals, showing excellent versatility.
The launch of FIORA not only fills the gap in mass spectrometry analysis, but also provides a powerful tool for future compound identification and research.