This repo provides the source code & data of our paper: QA-GNN: Reasoning with Language Models and Knowledge Graphs for Question Answering (NAACL 2021).
@InProceedings{yasunaga2021qagnn,
author = {Michihiro Yasunaga and Hongyu Ren and Antoine Bosselut and Percy Liang and Jure Leskovec},
title = {QA-GNN: Reasoning with Language Models and Knowledge Graphs for Question Answering},
year = {2021},
booktitle = {North American Chapter of the Association for Computational Linguistics (NAACL)},
}Webpage: https://snap.stanford.edu/qagnn
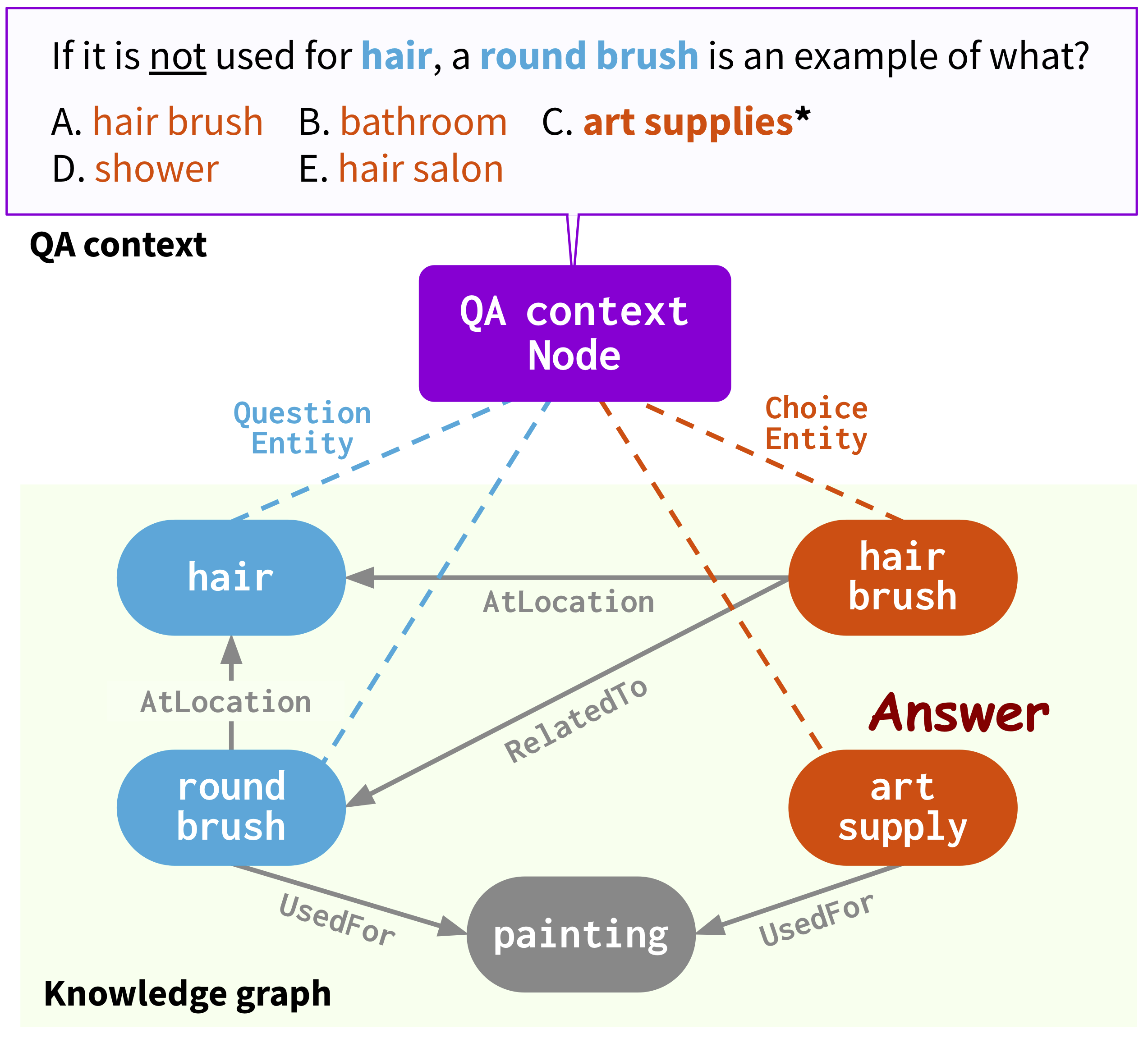
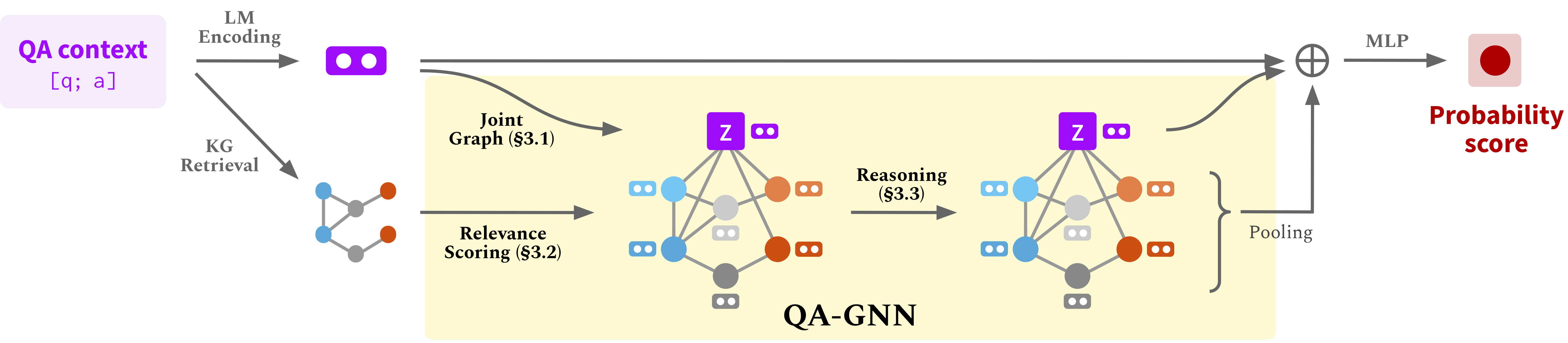
Run the following commands to create a conda environment (assuming CUDA10.1):
conda create -n qagnn python=3.7
source activate qagnn
pip install torch==1.8.0+cu101 -f https://download.pytorch.org/whl/torch_stable.html
pip install transformers==3.4.0
pip install nltk spacy==2.1.6
python -m spacy download en
# for torch-geometric
pip install torch-scatter==2.0.7 -f https://pytorch-geometric.com/whl/torch-1.8.0+cu101.html
pip install torch-sparse==0.6.9 -f https://pytorch-geometric.com/whl/torch-1.8.0+cu101.html
pip install torch-geometric==1.7.0 -f https://pytorch-geometric.com/whl/torch-1.8.0+cu101.htmlWe use the question answering datasets (CommonsenseQA, OpenBookQA) and the ConceptNet knowledge graph. Download all the raw data by
./download_raw_data.sh
Preprocess the raw data by running
python preprocess.py -p <num_processes>
The script will:
data/csqa/statement/)TL;DR (Skip above steps and just get preprocessed data). The preprocessing may take long. For your convenience, you can download all the processed data by
./download_preprocessed_data.sh
? NEWS (Add MedQA-USMLE). Besides the commonsense QA datasets (CommonsenseQA, OpenBookQA) with the ConceptNet knowledge graph, we added a biomedical QA dataset (MedQA-USMLE) with a biomedical knowledge graph based on Disease Database and DrugBank. You can download all the data for this from [here]. Unzip it and put the medqa_usmle and ddb folders inside the data/ directory. While this data is already preprocessed, we also provide the preprocessing scripts we used in utils_biomed/.
The resulting file structure will look like:
.
├── README.md
├── data/
├── cpnet/ (prerocessed ConceptNet)
├── csqa/
├── train_rand_split.jsonl
├── dev_rand_split.jsonl
├── test_rand_split_no_answers.jsonl
├── statement/ (converted statements)
├── grounded/ (grounded entities)
├── graphs/ (extracted subgraphs)
├── ...
├── obqa/
├── medqa_usmle/
└── ddb/
For CommonsenseQA, run
./run_qagnn__csqa.sh
For OpenBookQA, run
./run_qagnn__obqa.sh
For MedQA-USMLE, run
./run_qagnn__medqa_usmle.sh
As configured in these scripts, the model needs two types of input files
--{train,dev,test}_statements: preprocessed question statements in jsonl format. This is mainly loaded by load_input_tensors function in utils/data_utils.py.--{train,dev,test}_adj: information of the KG subgraph extracted for each question. This is mainly loaded by load_sparse_adj_data_with_contextnode function in utils/data_utils.py.Note: We find that training for OpenBookQA is unstable (e.g. best dev accuracy varies when using different seeds, different versions of the transformers / torch-geometric libraries, etc.), likely because the dataset is small. We suggest trying out different seeds. Another potential way to stabilize training is to initialize the model with one of the successful checkpoints provided below, e.g. by adding an argument --load_model_path obqa_model.pt.
For CommonsenseQA, run
./eval_qagnn__csqa.sh
Similarly, for other datasets (OpenBookQA, MedQA-USMLE), run ./eval_qagnn__obqa.sh and ./eval_qagnn__medqa_usmle.sh.
You can download trained model checkpoints in the next section.
CommonsenseQA
| Trained model | In-house Dev acc. | In-house Test acc. |
|---|---|---|
| RoBERTa-large + QA-GNN [link] | 0.7707 | 0.7405 |
OpenBookQA
| Trained model | Dev acc. | Test acc. |
|---|---|---|
| RoBERTa-large + QA-GNN [link] | 0.6960 | 0.6900 |
MedQA-USMLE
| Trained model | Dev acc. | Test acc. |
|---|---|---|
| SapBERT-base + QA-GNN [link] | 0.3789 | 0.3810 |
Note: The models were trained and tested with HuggingFace transformers==3.4.0.
{train,dev,test}.statement.jsonl in .jsonl format (see data/csqa/statement/train.statement.jsonl)data/{yourdataset}/ to store the .jsonl filespreprocess.py and perform subgraph extraction for your datautils/parser_utils.py to support your own datasetThis repo is built upon the following work:
Scalable Multi-Hop Relational Reasoning for Knowledge-Aware Question Answering. Yanlin Feng*, Xinyue Chen*, Bill Yuchen Lin, Peifeng Wang, Jun Yan and Xiang Ren. EMNLP 2020.
https://github.com/INK-USC/MHGRN
Many thanks to the authors and developers!